Gene
KWMTBOMO04899 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008018
Annotation
membrane_alanyl_aminopeptidase-like_[Bombyx_mori]
Full name
Aminopeptidase
Location in the cell
PlasmaMembrane Reliability : 2.922
Sequence
CDS
ATGGCCCGGAGAAAGTATAAGTGGAAATTCACTTTCTTCTATTTCTTGATACTATTGAACATTTTCACGAATTGTTTACCAATCGACAAGAAATTAAGGCAGACCACGAACGATGCTACATTCTCAGATTCGAATCCAGACGCAAACTCAGCTGAGTCCCACTTGTCCCAGCCGGATACTTTTGATGAAAAAACTAAGTTTATCCCAGAGGATGATTCGAAACGAACTGCGATTGTGTCAACTAATGCAGAAAGAAATCAGGCAGGAGAAACTGTCAGACCAGGTCAGACGATTAGTAGCTACGAAGTGGAAATCACCATAGAAGAAAATCGTTTCAAAGGAAGAGCCGTTATTGAATTGGAACTGGATGCCAATACGAGGGATGACCCCATAAAATTATATTCAGAGGATCTAGATGTAACCAGTGTACGGACCGGGGTCTTTACGGAAGCCAACAGCTTAGCAGCCACTTTCAGACTTAGAGATGGTTTTTTGGAGATCACTCCCGTACAGAGTACTACTTCATACGTTGTTGTGGTGGAATATACTGGAATACTAAGGAATGACGGCAACGGTATCTATCGCGGTGAAGGAAATGGCGTGTCATACCTGGCAATGAACCTACACCCTACACATGCGCGGCGCGTGTTTCCTTGCTTCGATGAACCTAACGAGGCAGCCAACATACGTTTCACGTTCAATGACGTCGAATTTAATAATATCGTCACAAATTCATTGCCCGAAGAAAATGCAGAGAATCGGTTCCGCGCCCTCGCCAGTTCTCCCCACAGGTGGGGAATGATCGCCCACGATTTCATTAATCTTAATATACCAACAACGGACGTGCTGTTGGTAGGACGACCGGGCCTCAGTAACCAAGATTCGCAAGCTTCATTAGCTATCAATGCATTCTTCAGCTTCCTGAACGAATGGACAGAAAAGCCTTACTTCGAGATCATTCTTAACCAGGAAAGTAGGATGCACGTTATCGCCTTACCCGATGTAGATCGTGACTGGCATGCTCTATCCATTGTTGGCATTTGGGAACCATATTTGTTGATGGAAACGACAGCTTCTGTTAAACAAAGAAAAACGGCGCTTGTGAAGATAGCAGAAACCATGGCGAAGCAGTGGTTTGGATACGTCATGTACCCCGAGAACTGGAGATTTGAATGGGTCTGTTAG
Protein
MARRKYKWKFTFFYFLILLNIFTNCLPIDKKLRQTTNDATFSDSNPDANSAESHLSQPDTFDEKTKFIPEDDSKRTAIVSTNAERNQAGETVRPGQTISSYEVEITIEENRFKGRAVIELELDANTRDDPIKLYSEDLDVTSVRTGVFTEANSLAATFRLRDGFLEITPVQSTTSYVVVVEYTGILRNDGNGIYRGEGNGVSYLAMNLHPTHARRVFPCFDEPNEAANIRFTFNDVEFNNIVTNSLPEENAENRFRALASSPHRWGMIAHDFINLNIPTTDVLLVGRPGLSNQDSQASLAINAFFSFLNEWTEKPYFEIILNQESRMHVIALPDVDRDWHALSIVGIWEPYLLMETTASVKQRKTALVKIAETMAKQWFGYVMYPENWRFEWVC
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Feature
chain Aminopeptidase
Uniprot
I3VR82
H9JES1
A0A194QR13
A0A2W1BM20
A0A194QD33
A0A2H1VVS9
+ More
A0A212EJS2 A0A286LQ00 A0A2A4J290 A0A2H1VG23 A0A0L7LHA5 E2BB80 A0A3A4VBH3 D6WBB0 A0A1F6V3Z0 Q16ZL6 A0A017TCZ3 K7INE4 A0A1M3N3Y2 A0A0P6ET91 A0A232EWK1 A0A1F6Y568 A0A1F5JQ82 A0A1S4FIM9 A0A2H0UB43 A0A0G1GT22 A0A352BEA5 A0A0T6BAA6 A0A1F5KPK5 A0A182H3I7 A0A2P6N3L2 A0A0G0QRZ6 A0A2W0AY25 W7L7A4 A0A182HSV0 A0A2W0C125 A0A3Q3BI68 A0A210PQ87 A0A2E6LF19 Q7QC91 A0A0P5QDI9 A0A0P5PZM3 A0A0P5DDZ9 A0A1F6WY42 A0A0F2LTP9 A0A0P5CKV4 A0A1J1IMJ3 A0A0P6A6Q7 A0A0P5VUF0 A0A0P5K4F5 A0A0P6FNJ7 A0A2A3ELA7 A0A0N8BG95 A0A0P5D0N1 A0A0P5LKV9 A0A0P6BZ76 A0A0P5MJ75 A0A0P5MJN9 A0A0P6G6P9 A0A0P5ZUC5 A0A0N8AXC2
A0A212EJS2 A0A286LQ00 A0A2A4J290 A0A2H1VG23 A0A0L7LHA5 E2BB80 A0A3A4VBH3 D6WBB0 A0A1F6V3Z0 Q16ZL6 A0A017TCZ3 K7INE4 A0A1M3N3Y2 A0A0P6ET91 A0A232EWK1 A0A1F6Y568 A0A1F5JQ82 A0A1S4FIM9 A0A2H0UB43 A0A0G1GT22 A0A352BEA5 A0A0T6BAA6 A0A1F5KPK5 A0A182H3I7 A0A2P6N3L2 A0A0G0QRZ6 A0A2W0AY25 W7L7A4 A0A182HSV0 A0A2W0C125 A0A3Q3BI68 A0A210PQ87 A0A2E6LF19 Q7QC91 A0A0P5QDI9 A0A0P5PZM3 A0A0P5DDZ9 A0A1F6WY42 A0A0F2LTP9 A0A0P5CKV4 A0A1J1IMJ3 A0A0P6A6Q7 A0A0P5VUF0 A0A0P5K4F5 A0A0P6FNJ7 A0A2A3ELA7 A0A0N8BG95 A0A0P5D0N1 A0A0P5LKV9 A0A0P6BZ76 A0A0P5MJ75 A0A0P5MJN9 A0A0P6G6P9 A0A0P5ZUC5 A0A0N8AXC2
EC Number
3.4.11.-
Pubmed
EMBL
JQ061153
AFK85027.1
BABH01023395
BABH01023396
BABH01023397
BABH01023398
+ More
KQ461198 KPJ05971.1 KZ149969 PZC76122.1 KQ459185 KPJ03372.1 ODYU01004747 SOQ44951.1 AGBW02014405 OWR41739.1 MF425657 ASU92549.1 NWSH01003448 PCG66277.1 ODYU01002173 SOQ39332.1 JTDY01001142 KOB74749.1 GL446946 EFN87053.1 QZIY01000019 RJQ34644.1 KQ971312 EEZ99298.1 MFTO01000001 OGI64393.1 CH477489 EAT40058.1 ASRX01000014 EYF06797.1 MKVQ01000024 OJY23046.1 GDIQ01071637 JAN23100.1 NNAY01001852 OXU22726.1 MFVU01000024 OGJ01507.1 MFCV01000044 OGE30746.1 PFBK01000008 PIR83629.1 LCHR01000017 KKT37403.1 DNMN01000018 HBB44037.1 LJIG01002688 KRT84252.1 MFDM01000020 OGE42848.1 JXUM01003831 KQ560164 KXJ84068.1 MDYQ01000218 PRP78523.1 LBXY01000002 KKR40106.1 QIBB01000497 PYY10602.1 ASRH01000003 EWG07574.1 APCN01000366 QIBC01000015 PYY19931.1 NEDP02005559 OWF38669.1 PBJQ01000011 MBK77236.1 AAAB01008859 EAA08230.5 GDIQ01121870 JAL29856.1 GDIQ01121869 JAL29857.1 GDIP01162826 JAJ60576.1 MFUR01000010 OGI86809.1 JZWS01000017 KJR79196.1 GDIP01169022 JAJ54380.1 CVRI01000055 CRL01384.1 GDIP01035094 LRGB01002121 JAM68621.1 KZS09066.1 GDIP01109686 JAL94028.1 GDIQ01194840 JAK56885.1 GDIQ01046749 JAN47988.1 KZ288223 PBC31976.1 GDIQ01180699 JAK71026.1 GDIP01162827 JAJ60575.1 GDIQ01193049 JAK58676.1 GDIP01007169 GDIP01007168 GDIQ01035004 JAM96546.1 JAN59733.1 GDIQ01155913 JAK95812.1 GDIQ01162754 JAK88971.1 GDIQ01048296 JAN46441.1 GDIP01039699 JAM64016.1 GDIQ01233683 JAK18042.1
KQ461198 KPJ05971.1 KZ149969 PZC76122.1 KQ459185 KPJ03372.1 ODYU01004747 SOQ44951.1 AGBW02014405 OWR41739.1 MF425657 ASU92549.1 NWSH01003448 PCG66277.1 ODYU01002173 SOQ39332.1 JTDY01001142 KOB74749.1 GL446946 EFN87053.1 QZIY01000019 RJQ34644.1 KQ971312 EEZ99298.1 MFTO01000001 OGI64393.1 CH477489 EAT40058.1 ASRX01000014 EYF06797.1 MKVQ01000024 OJY23046.1 GDIQ01071637 JAN23100.1 NNAY01001852 OXU22726.1 MFVU01000024 OGJ01507.1 MFCV01000044 OGE30746.1 PFBK01000008 PIR83629.1 LCHR01000017 KKT37403.1 DNMN01000018 HBB44037.1 LJIG01002688 KRT84252.1 MFDM01000020 OGE42848.1 JXUM01003831 KQ560164 KXJ84068.1 MDYQ01000218 PRP78523.1 LBXY01000002 KKR40106.1 QIBB01000497 PYY10602.1 ASRH01000003 EWG07574.1 APCN01000366 QIBC01000015 PYY19931.1 NEDP02005559 OWF38669.1 PBJQ01000011 MBK77236.1 AAAB01008859 EAA08230.5 GDIQ01121870 JAL29856.1 GDIQ01121869 JAL29857.1 GDIP01162826 JAJ60576.1 MFUR01000010 OGI86809.1 JZWS01000017 KJR79196.1 GDIP01169022 JAJ54380.1 CVRI01000055 CRL01384.1 GDIP01035094 LRGB01002121 JAM68621.1 KZS09066.1 GDIP01109686 JAL94028.1 GDIQ01194840 JAK56885.1 GDIQ01046749 JAN47988.1 KZ288223 PBC31976.1 GDIQ01180699 JAK71026.1 GDIP01162827 JAJ60575.1 GDIQ01193049 JAK58676.1 GDIP01007169 GDIP01007168 GDIQ01035004 JAM96546.1 JAN59733.1 GDIQ01155913 JAK95812.1 GDIQ01162754 JAK88971.1 GDIQ01048296 JAN46441.1 GDIP01039699 JAM64016.1 GDIQ01233683 JAK18042.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
I3VR82
H9JES1
A0A194QR13
A0A2W1BM20
A0A194QD33
A0A2H1VVS9
+ More
A0A212EJS2 A0A286LQ00 A0A2A4J290 A0A2H1VG23 A0A0L7LHA5 E2BB80 A0A3A4VBH3 D6WBB0 A0A1F6V3Z0 Q16ZL6 A0A017TCZ3 K7INE4 A0A1M3N3Y2 A0A0P6ET91 A0A232EWK1 A0A1F6Y568 A0A1F5JQ82 A0A1S4FIM9 A0A2H0UB43 A0A0G1GT22 A0A352BEA5 A0A0T6BAA6 A0A1F5KPK5 A0A182H3I7 A0A2P6N3L2 A0A0G0QRZ6 A0A2W0AY25 W7L7A4 A0A182HSV0 A0A2W0C125 A0A3Q3BI68 A0A210PQ87 A0A2E6LF19 Q7QC91 A0A0P5QDI9 A0A0P5PZM3 A0A0P5DDZ9 A0A1F6WY42 A0A0F2LTP9 A0A0P5CKV4 A0A1J1IMJ3 A0A0P6A6Q7 A0A0P5VUF0 A0A0P5K4F5 A0A0P6FNJ7 A0A2A3ELA7 A0A0N8BG95 A0A0P5D0N1 A0A0P5LKV9 A0A0P6BZ76 A0A0P5MJ75 A0A0P5MJN9 A0A0P6G6P9 A0A0P5ZUC5 A0A0N8AXC2
A0A212EJS2 A0A286LQ00 A0A2A4J290 A0A2H1VG23 A0A0L7LHA5 E2BB80 A0A3A4VBH3 D6WBB0 A0A1F6V3Z0 Q16ZL6 A0A017TCZ3 K7INE4 A0A1M3N3Y2 A0A0P6ET91 A0A232EWK1 A0A1F6Y568 A0A1F5JQ82 A0A1S4FIM9 A0A2H0UB43 A0A0G1GT22 A0A352BEA5 A0A0T6BAA6 A0A1F5KPK5 A0A182H3I7 A0A2P6N3L2 A0A0G0QRZ6 A0A2W0AY25 W7L7A4 A0A182HSV0 A0A2W0C125 A0A3Q3BI68 A0A210PQ87 A0A2E6LF19 Q7QC91 A0A0P5QDI9 A0A0P5PZM3 A0A0P5DDZ9 A0A1F6WY42 A0A0F2LTP9 A0A0P5CKV4 A0A1J1IMJ3 A0A0P6A6Q7 A0A0P5VUF0 A0A0P5K4F5 A0A0P6FNJ7 A0A2A3ELA7 A0A0N8BG95 A0A0P5D0N1 A0A0P5LKV9 A0A0P6BZ76 A0A0P5MJ75 A0A0P5MJN9 A0A0P6G6P9 A0A0P5ZUC5 A0A0N8AXC2
PDB
5CU5
E-value=2.20096e-07,
Score=132
Ontologies
PATHWAY
GO
PANTHER
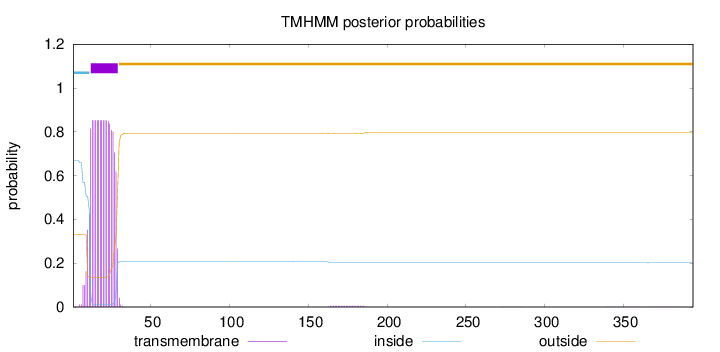
Topology
SignalP
Position: 1 - 25,
Likelihood: 0.825796
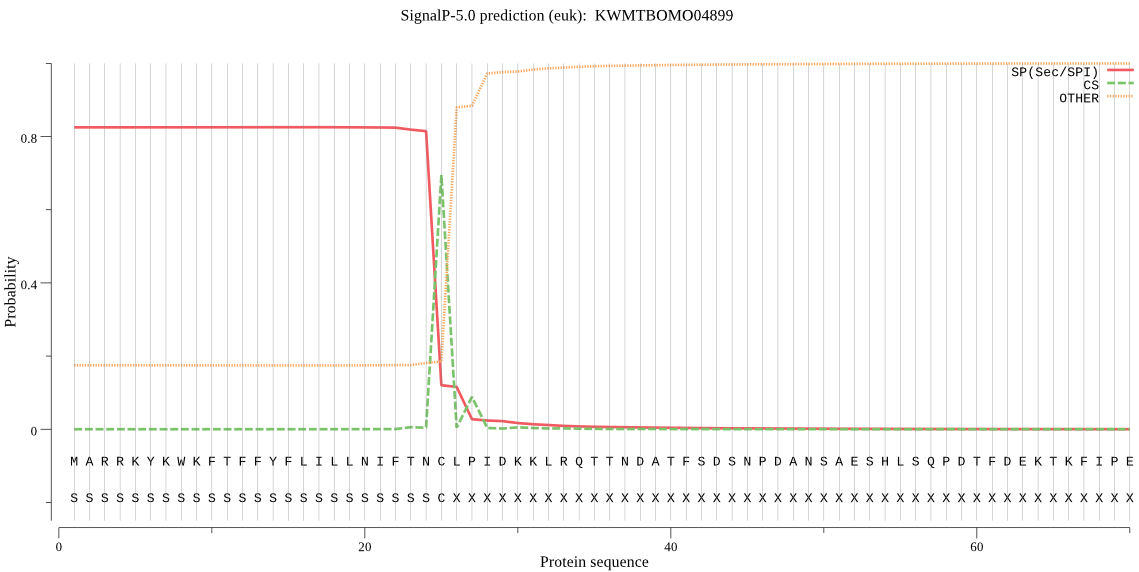
Length:
394
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.58663
Exp number, first 60 AAs:
15.43842
Total prob of N-in:
0.66942
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 29
outside
30 - 394
Population Genetic Test Statistics
Pi
176.882867
Theta
58.71478
Tajima's D
0.887958
CLR
22.145831
CSRT
0.630668466576671
Interpretation
Uncertain
