Gene
KWMTBOMO04895 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008018
Annotation
membrane_alanyl_aminopeptidase-like_[Bombyx_mori]
Full name
Aminopeptidase
Sequence
CDS
ATGGTGCGTAGAACTGTGGGTGCACTTGAAGACGAAATTGGATTCAATCCTGATACATCACTCACTGAGCCTGCAATGGTTTCCTTGACCAGAGGTCTGGTAATGGATCACGCCTGCAGGGCCAATTATGAGCCATGCATTGCAGCCGCTATCGATTTGTTCTATAATCCCAATAACAATGAAGAAGTAAATCCCAACATCCCATCTGATATCAGACCAGCGGTCTACTGTACCATGGTAAGAGAAGGAGATGAGGACGTGATAGAAGCATTGAAAGCTAGATTGGAAGTTGAGGCAAACCATTACGAGCGAGTTGTCATATTAGAGTCGTTGGCCTGCTCAGATGATCAAAACTTTATACGAAATTACCTTGAAGAAACGATAGCTGCAGGTAATGAATACGGTGTGGAAGAAAGAGTCAGGATATTTAGAGCAGTAGCTGAAAGTAGTTACGAGAATGCGAGAGTGGCTTTAAGCTTTATTTCAATGCGCACTAATGAGATAAGAGATAATTATGGCGGCCCGAAAAAATTAGAAGAGATACTTTTCGTATTGGGTGAAAACATGGCGAATGATATATTATCTGAAGACTTCCGGATTTGGGTGAGATCGCAGTCAAATGATTTGGATGATTCTCAAGGAGCTGCAAACAGGGCATTGGCGATTGTTCTTGAGAATGTCAACTGGATTGAGCGCCACCAAGATTATGTCTACGATTGGATAGACGAGAACGACGCACCAACTTTAGCTGCATCATTAATTCTAATTTGTATAACAATCTTTGTTACTCTATTTAATAATTAG
Protein
MVRRTVGALEDEIGFNPDTSLTEPAMVSLTRGLVMDHACRANYEPCIAAAIDLFYNPNNNEEVNPNIPSDIRPAVYCTMVREGDEDVIEALKARLEVEANHYERVVILESLACSDDQNFIRNYLEETIAAGNEYGVEERVRIFRAVAESSYENARVALSFISMRTNEIRDNYGGPKKLEEILFVLGENMANDILSEDFRIWVRSQSNDLDDSQGAANRALAIVLENVNWIERHQDYVYDWIDENDAPTLAASLILICITIFVTLFNN
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Feature
chain Aminopeptidase
Uniprot
I3VR82
H9JES1
A0A194QR13
A0A194QD33
A0A286LQ00
A0A2H1VG23
+ More
A0A2W1BM20 A0A0L7LHA5 A0A2H1VVS9 A0A2A4IVH9 A0A2A4J290 A0A212EJS2 Q8MUT5 Q7Z268 Q8MU79 A0A2W1BSE9 Q9NHZ8 Q86QI6 C9W8K9 Q963G0 Q0PDN2 A0A0L7L425 F2YWM1 G9C5G4 A0A2J7RAG9 A0A2J7RAH0 A0A182XHW8 A0A087ZUB2 A0A1L8E2T9 A4D0J6 A0A2H1WAU1 A0A067RF43 Q95WL1 A0A1B0CGE2 A0A182HSV0 Q7QC91 A0A2A4J832 Q964F9 A0A182PUT5 A0A212EJS8 A0A310SKF7 A0A2P8Y405 Q9XYV3 G9C5G3 A0A2J7RAG5 A0A0C9R4H9 A0A0N0BIW4 A0A0T6B4M8 A0A1W5BDR3 A0A1W5BEF8 F6X9G8 B3P5I5 Q4G6A2 T1P9B0 A0A0C9RIL7 A0A076YIK8 A0A194QD38 A0A1I8N5N4 U5EWR8 A0A087SVG3 A0A154PML5 Q6PWP5 F4W4E4 A0A212EJU3 O46156 A0A158P3F5 Q6J4J3 G3ESV7 D3YJ03 A0A0C9RIM2 S4VDN0 A0A026VYW9 A0A0C9QEZ7 K7J1L8 A0A3L8E5F4 D2KHN2 A0A194QR18 A0A194QK79 A0A232FGY1 A0A2R7VTS0 O77046 A0A1Q3FS18 D2KHN1 D3YJ04 A0A2Z3YIN5 A0A087ZUB4 A0A1W4VR30 B2LS41 Q8T5Q4 G9C7N4 Q6Q2I1 A0A1I9WL50 A0A2L1IQ82 A0A182W7Y5 Q9VAM2 A0A1S5QN38 A0A2A3ELA7
A0A2W1BM20 A0A0L7LHA5 A0A2H1VVS9 A0A2A4IVH9 A0A2A4J290 A0A212EJS2 Q8MUT5 Q7Z268 Q8MU79 A0A2W1BSE9 Q9NHZ8 Q86QI6 C9W8K9 Q963G0 Q0PDN2 A0A0L7L425 F2YWM1 G9C5G4 A0A2J7RAG9 A0A2J7RAH0 A0A182XHW8 A0A087ZUB2 A0A1L8E2T9 A4D0J6 A0A2H1WAU1 A0A067RF43 Q95WL1 A0A1B0CGE2 A0A182HSV0 Q7QC91 A0A2A4J832 Q964F9 A0A182PUT5 A0A212EJS8 A0A310SKF7 A0A2P8Y405 Q9XYV3 G9C5G3 A0A2J7RAG5 A0A0C9R4H9 A0A0N0BIW4 A0A0T6B4M8 A0A1W5BDR3 A0A1W5BEF8 F6X9G8 B3P5I5 Q4G6A2 T1P9B0 A0A0C9RIL7 A0A076YIK8 A0A194QD38 A0A1I8N5N4 U5EWR8 A0A087SVG3 A0A154PML5 Q6PWP5 F4W4E4 A0A212EJU3 O46156 A0A158P3F5 Q6J4J3 G3ESV7 D3YJ03 A0A0C9RIM2 S4VDN0 A0A026VYW9 A0A0C9QEZ7 K7J1L8 A0A3L8E5F4 D2KHN2 A0A194QR18 A0A194QK79 A0A232FGY1 A0A2R7VTS0 O77046 A0A1Q3FS18 D2KHN1 D3YJ04 A0A2Z3YIN5 A0A087ZUB4 A0A1W4VR30 B2LS41 Q8T5Q4 G9C7N4 Q6Q2I1 A0A1I9WL50 A0A2L1IQ82 A0A182W7Y5 Q9VAM2 A0A1S5QN38 A0A2A3ELA7
EC Number
3.4.11.-
Pubmed
19121390
26354079
28756777
26227816
22118469
11439249
+ More
20824821 17402938 15857766 24845553 12364791 14747013 17210077 12200317 29403074 10406091 12481130 15114417 17994087 15978131 25315136 21719571 21347285 24121099 24508170 20075255 30249741 28648823 27538518 29415259 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
20824821 17402938 15857766 24845553 12364791 14747013 17210077 12200317 29403074 10406091 12481130 15114417 17994087 15978131 25315136 21719571 21347285 24121099 24508170 20075255 30249741 28648823 27538518 29415259 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
EMBL
JQ061153
AFK85027.1
BABH01023395
BABH01023396
BABH01023397
BABH01023398
+ More
KQ461198 KPJ05971.1 KQ459185 KPJ03372.1 MF425657 ASU92549.1 ODYU01002173 SOQ39332.1 KZ149969 PZC76122.1 JTDY01001142 KOB74749.1 ODYU01004747 SOQ44951.1 NWSH01006029 PCG63731.1 NWSH01003448 PCG66277.1 AGBW02014405 OWR41739.1 AF511038 AAM44056.1 AY279534 AAP37950.1 AF535165 AAN04899.1 PZC76117.1 AF217249 AAF37559.1 AY181026 AAO23562.1 FJ210822 ACN69219.1 AF378666 AAK58066.1 DQ872666 ABH07377.2 JTDY01003068 KOB70212.1 JF509138 ADZ74247.1 HQ853296 ADZ05468.1 NEVH01006570 PNF37826.1 PNF37825.1 GFDF01001118 JAV12966.1 AY836582 AAX39866.1 ODYU01007373 SOQ50076.1 KK852505 KDR22501.1 AF317619 AAL26894.1 AJWK01011046 AJWK01011047 APCN01000366 AAAB01008859 EAA08230.5 NWSH01002538 PCG68029.1 AF320764 AAK69605.1 OWR41736.1 KQ760397 OAD60664.1 PYGN01000952 PSN39002.1 AF126443 AAD31184.1 HQ853295 ADZ05467.1 PNF37829.1 GBYB01002810 GBYB01007837 JAG72577.1 JAG77604.1 KQ435724 KOX78331.1 LJIG01009863 KRT82230.1 EAAA01002576 CH954182 EDV53235.1 AY218845 AAP44967.1 KA644745 AFP59374.1 GBYB01013077 JAG82844.1 KM034756 AIK67332.1 KPJ03377.1 GANO01001287 JAB58584.1 KK112145 KFM56852.1 KQ434984 KZC13131.1 AY573579 AAS82738.1 GL887532 EGI70839.1 OWR41740.1 AJ222699 CAA10950.1 ADTU01007783 ADTU01007784 ADTU01007785 AY601813 AAT45356.1 JF339039 AEO12694.1 GU479676 ADD39717.1 GBYB01013082 JAG82849.1 KC776603 AGO57907.1 KK107589 EZA48631.1 GBYB01013078 GBYB01013080 JAG82845.1 JAG82847.1 QOIP01000001 RLU27565.1 GU213037 ADA59489.1 KPJ05976.1 KPJ05978.1 NNAY01000207 OXU30026.1 KK854013 PTY09335.1 AB013400 BAA33715.1 GFDL01004812 JAV30233.1 GU213036 ADA59488.1 GU479677 ADD39718.1 KY949254 AWT22996.1 EU571948 ACB87202.2 AF498996 AAM18718.1 HQ901596 ADZ57273.1 AY564236 AAS75551.1 KU932234 APA33870.1 MF741655 AVD96938.1 AE014297 AAF56882.2 KR068519 AMO02536.1 KZ288223 PBC31976.1
KQ461198 KPJ05971.1 KQ459185 KPJ03372.1 MF425657 ASU92549.1 ODYU01002173 SOQ39332.1 KZ149969 PZC76122.1 JTDY01001142 KOB74749.1 ODYU01004747 SOQ44951.1 NWSH01006029 PCG63731.1 NWSH01003448 PCG66277.1 AGBW02014405 OWR41739.1 AF511038 AAM44056.1 AY279534 AAP37950.1 AF535165 AAN04899.1 PZC76117.1 AF217249 AAF37559.1 AY181026 AAO23562.1 FJ210822 ACN69219.1 AF378666 AAK58066.1 DQ872666 ABH07377.2 JTDY01003068 KOB70212.1 JF509138 ADZ74247.1 HQ853296 ADZ05468.1 NEVH01006570 PNF37826.1 PNF37825.1 GFDF01001118 JAV12966.1 AY836582 AAX39866.1 ODYU01007373 SOQ50076.1 KK852505 KDR22501.1 AF317619 AAL26894.1 AJWK01011046 AJWK01011047 APCN01000366 AAAB01008859 EAA08230.5 NWSH01002538 PCG68029.1 AF320764 AAK69605.1 OWR41736.1 KQ760397 OAD60664.1 PYGN01000952 PSN39002.1 AF126443 AAD31184.1 HQ853295 ADZ05467.1 PNF37829.1 GBYB01002810 GBYB01007837 JAG72577.1 JAG77604.1 KQ435724 KOX78331.1 LJIG01009863 KRT82230.1 EAAA01002576 CH954182 EDV53235.1 AY218845 AAP44967.1 KA644745 AFP59374.1 GBYB01013077 JAG82844.1 KM034756 AIK67332.1 KPJ03377.1 GANO01001287 JAB58584.1 KK112145 KFM56852.1 KQ434984 KZC13131.1 AY573579 AAS82738.1 GL887532 EGI70839.1 OWR41740.1 AJ222699 CAA10950.1 ADTU01007783 ADTU01007784 ADTU01007785 AY601813 AAT45356.1 JF339039 AEO12694.1 GU479676 ADD39717.1 GBYB01013082 JAG82849.1 KC776603 AGO57907.1 KK107589 EZA48631.1 GBYB01013078 GBYB01013080 JAG82845.1 JAG82847.1 QOIP01000001 RLU27565.1 GU213037 ADA59489.1 KPJ05976.1 KPJ05978.1 NNAY01000207 OXU30026.1 KK854013 PTY09335.1 AB013400 BAA33715.1 GFDL01004812 JAV30233.1 GU213036 ADA59488.1 GU479677 ADD39718.1 KY949254 AWT22996.1 EU571948 ACB87202.2 AF498996 AAM18718.1 HQ901596 ADZ57273.1 AY564236 AAS75551.1 KU932234 APA33870.1 MF741655 AVD96938.1 AE014297 AAF56882.2 KR068519 AMO02536.1 KZ288223 PBC31976.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000218220
UP000007151
+ More
UP000235965 UP000076407 UP000005203 UP000027135 UP000092461 UP000075840 UP000007062 UP000075885 UP000245037 UP000053105 UP000008144 UP000008711 UP000095301 UP000054359 UP000076502 UP000007755 UP000005205 UP000053097 UP000002358 UP000279307 UP000215335 UP000192221 UP000075920 UP000000803 UP000242457
UP000235965 UP000076407 UP000005203 UP000027135 UP000092461 UP000075840 UP000007062 UP000075885 UP000245037 UP000053105 UP000008144 UP000008711 UP000095301 UP000054359 UP000076502 UP000007755 UP000005205 UP000053097 UP000002358 UP000279307 UP000215335 UP000192221 UP000075920 UP000000803 UP000242457
Interpro
SUPFAM
SSF48350
SSF48350
Gene 3D
CDD
ProteinModelPortal
I3VR82
H9JES1
A0A194QR13
A0A194QD33
A0A286LQ00
A0A2H1VG23
+ More
A0A2W1BM20 A0A0L7LHA5 A0A2H1VVS9 A0A2A4IVH9 A0A2A4J290 A0A212EJS2 Q8MUT5 Q7Z268 Q8MU79 A0A2W1BSE9 Q9NHZ8 Q86QI6 C9W8K9 Q963G0 Q0PDN2 A0A0L7L425 F2YWM1 G9C5G4 A0A2J7RAG9 A0A2J7RAH0 A0A182XHW8 A0A087ZUB2 A0A1L8E2T9 A4D0J6 A0A2H1WAU1 A0A067RF43 Q95WL1 A0A1B0CGE2 A0A182HSV0 Q7QC91 A0A2A4J832 Q964F9 A0A182PUT5 A0A212EJS8 A0A310SKF7 A0A2P8Y405 Q9XYV3 G9C5G3 A0A2J7RAG5 A0A0C9R4H9 A0A0N0BIW4 A0A0T6B4M8 A0A1W5BDR3 A0A1W5BEF8 F6X9G8 B3P5I5 Q4G6A2 T1P9B0 A0A0C9RIL7 A0A076YIK8 A0A194QD38 A0A1I8N5N4 U5EWR8 A0A087SVG3 A0A154PML5 Q6PWP5 F4W4E4 A0A212EJU3 O46156 A0A158P3F5 Q6J4J3 G3ESV7 D3YJ03 A0A0C9RIM2 S4VDN0 A0A026VYW9 A0A0C9QEZ7 K7J1L8 A0A3L8E5F4 D2KHN2 A0A194QR18 A0A194QK79 A0A232FGY1 A0A2R7VTS0 O77046 A0A1Q3FS18 D2KHN1 D3YJ04 A0A2Z3YIN5 A0A087ZUB4 A0A1W4VR30 B2LS41 Q8T5Q4 G9C7N4 Q6Q2I1 A0A1I9WL50 A0A2L1IQ82 A0A182W7Y5 Q9VAM2 A0A1S5QN38 A0A2A3ELA7
A0A2W1BM20 A0A0L7LHA5 A0A2H1VVS9 A0A2A4IVH9 A0A2A4J290 A0A212EJS2 Q8MUT5 Q7Z268 Q8MU79 A0A2W1BSE9 Q9NHZ8 Q86QI6 C9W8K9 Q963G0 Q0PDN2 A0A0L7L425 F2YWM1 G9C5G4 A0A2J7RAG9 A0A2J7RAH0 A0A182XHW8 A0A087ZUB2 A0A1L8E2T9 A4D0J6 A0A2H1WAU1 A0A067RF43 Q95WL1 A0A1B0CGE2 A0A182HSV0 Q7QC91 A0A2A4J832 Q964F9 A0A182PUT5 A0A212EJS8 A0A310SKF7 A0A2P8Y405 Q9XYV3 G9C5G3 A0A2J7RAG5 A0A0C9R4H9 A0A0N0BIW4 A0A0T6B4M8 A0A1W5BDR3 A0A1W5BEF8 F6X9G8 B3P5I5 Q4G6A2 T1P9B0 A0A0C9RIL7 A0A076YIK8 A0A194QD38 A0A1I8N5N4 U5EWR8 A0A087SVG3 A0A154PML5 Q6PWP5 F4W4E4 A0A212EJU3 O46156 A0A158P3F5 Q6J4J3 G3ESV7 D3YJ03 A0A0C9RIM2 S4VDN0 A0A026VYW9 A0A0C9QEZ7 K7J1L8 A0A3L8E5F4 D2KHN2 A0A194QR18 A0A194QK79 A0A232FGY1 A0A2R7VTS0 O77046 A0A1Q3FS18 D2KHN1 D3YJ04 A0A2Z3YIN5 A0A087ZUB4 A0A1W4VR30 B2LS41 Q8T5Q4 G9C7N4 Q6Q2I1 A0A1I9WL50 A0A2L1IQ82 A0A182W7Y5 Q9VAM2 A0A1S5QN38 A0A2A3ELA7
PDB
4WZ9
E-value=5.81276e-05,
Score=109
Ontologies
GO
PANTHER
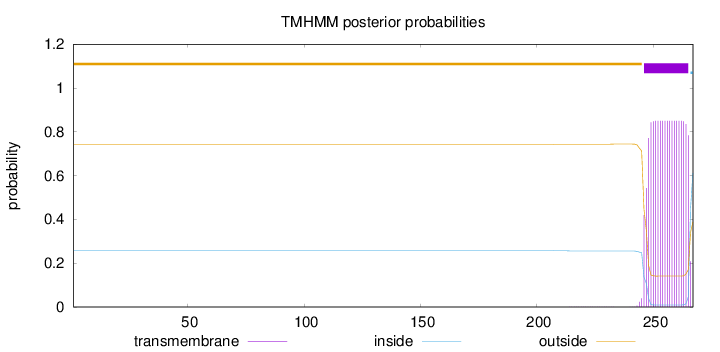
Topology
Length:
267
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
16.3911
Exp number, first 60 AAs:
0.00146
Total prob of N-in:
0.25683
outside
1 - 245
TMhelix
246 - 265
inside
266 - 267
Population Genetic Test Statistics
Pi
160.451739
Theta
156.5994
Tajima's D
-0.335949
CLR
1.006651
CSRT
0.28163591820409
Interpretation
Uncertain
