Gene
KWMTBOMO04894 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008063
Annotation
glutamyl_aminopeptidase-like_precursor_[Bombyx_mori]
Full name
Aminopeptidase
Location in the cell
PlasmaMembrane Reliability : 3.949
Sequence
CDS
ATGTTTTTCGCATTGTTATTGTTCGTTCTCTTTTCATATTCGAACGCGAATTTTATCGTTGAAGAAGAATGCTTGAATTACACGGTTTATCCTATACAATACGAACTGACAATCATACCTTATATTTACAAAGACAATTCCTATTACCACGGCGATATAACGATCACTGTTATTGCGAACGCCAATGTAAGGGAGATCGAGTTAGACGCGAAGGATTTAGACATCCAGAGCGGTTCCATCAAGGTCTTGGACGGAAGCACTGATCTGGTGAATGGAGCACGTCCGTATGAATACGACAAGACTAATGGGAAGCTGTTCATTCACTTGCGGGAGCCTTTGAAGGTGTACAGTCAAAACAATAGACAGTTCTACTATATAAAAATGTCTTTTAATAAATACATCAAAGAAGATAGCGCCGGTTTGTTCTTGGTCAACTATTATGAGGACGATGTGAAGAATGCCAAGTCTTTATACGCAACCAGGCTGTCTCCGAATAAGGCTAAATTCATGTTTCCGTGCTTCGATAACCCTCGCTTCGAAGCGGTCTTCAAATTCAAGGTGTACATACTTTCCGATCACCCAGGAATGCAGTACACTAATAGCAGCTTGGTGATCGCTGAGGAAATGAAGAGAGTCAGCTCTAAAGACTATACGATCATCGAATACATTCCATCACCTCAAGTAGCATTGCACCAAGTCGGGTTTCACTATTCCCAATTCACCAATAAATCGGTCAAAGGCAAAAATAATGATACCCTTGTTATCTGGGCACCAGGCGATAAATTGTATTGCTATGATTACATACAACGCTTTGGAATATCCATGATCGATCTCATGCATCAGTATGAGGGTACCAAGAGACCTATAACTGCGGGGAGTATCAATTTAGTTGCAGTTCCCACTAATTTACAAGGATATGAGATTGGCAGTTGGAATCTACTGACTAATGGTGCTAACCGCATAGCAAATCTGGTGTATATAACAAGTATCCAGCAAATAGAAGTGATGCGGTTCGAATTAGCCCAGCAGTTGTGCAGGATTTGGCTTGGTAATCCTGGTGAAGAGAACCGCACCAGGTGGAAGGAGGAATGGCTGAAGGAAGGAATGGCTACTTACTATGCTTCTTATTTTCTGGCTCAGAAAACATCAGAGCGTTATCCAAAAAGACTACAACCATTAGGTATATACGGATTGTCCATGAAACATAAAGCCATGGAAATCGACTGGCCCAGAAGTACTCCAGCATTGGAAGTTTTCAATGACACATTGGCCATAGAAATACCAGCTAGATACAAGGATTTGGTCACTATGAAAACTGGGACGCTTTTCTGGATGTTAGAAAATTGGATTGGACCAGAGAAATTTCATAATGCCTTAATCAAATACATTAATTCAAGAAGAGGCAAGTACATATCCCTTCAAGATTTCCTGACGACGTTGGACCACGAGACGGTGGAATGCTCGTATCAGCTCTTCAACGGTTCCACGTCATCCAGGATACTCAGGTCTTGGTTCAGTCAGGCTGGATACCCTGTTATTAATGTGAGAGTTCTCAGAGATCGAACACCAAACGTTGTTCAACTGAATCAAAGGAGATTCAGCTTCAGGAATGAACACCATCGAGAGTTGGAGTACCTTATTCCCATTTCCTACACGATCCAGAACAGCAAGAACTGTTTCAATTGCTACAGGCCTCGTTTCGTTATTGGAAGTCAACCGTACACATTCATCGAAAACCTGGATGGGGGCTGGATTATCCTAAACAGCAATGGCTCAGGTTACTATCGCGTGAATTATGACTCTGACAGTTGGTTGCTGATTGCCAAGACTCTTGTCGAAGATCATCAATCTATTGAAGAAATGACTAGATCTCAAATAGTCAACGATGTATTCGCTCTGTATGTGTCTGGGAACTTGGACATGTCTATAGCTATGCAGGTTTTGGCGTACCTCGATAAAGAGACCAGTCTTGCTGTATGGGACAGCGTAATATCGGGGTACGAGCTGCTGAAGCTGGAAGGAGCAGCTTGTAATATGACCAAGTACATATATTGGGAATGGTCGGATTTCATGAAGCAGAAGATACCGACTATATACAATGGCCTAATTAAGAATCTCGATCAGCAAACAGATGTAAGGCTGCTCCGCAACAAAGTCATTACATTCGCAGCTGAACTAGAATATGAACAGTGCTTGACACAGTTGCGCCGGATGTATCAGGAGACGCGCAGAGATCCAAAACGCATTCTCGATCCAGACGCAAGACGTGGATGTTACTACGTCCTTTTGAACGACACAAACAGTCACCTCCGGTTCAAAAAACTTAGCGGTATCGAGGAAGAAGACCGGAATTCCGTTGAAAACAGAATACGCGAAAATAACAGATTCCTCATCAGGATACCGTACGGAGAACCCAGACCGTTGCCTATTATATCATCTGAAAGTCCATTATCTTCGACTACGCCTGAGCCGGTACCAGACAAGATCGTAATGCCCGACGCGTCCACAAGCAACTTATCTTCTATAATATTTGTAGTCGCTTCAATACTGATACTGAGGCTGTTTTGA
Protein
MFFALLLFVLFSYSNANFIVEEECLNYTVYPIQYELTIIPYIYKDNSYYHGDITITVIANANVREIELDAKDLDIQSGSIKVLDGSTDLVNGARPYEYDKTNGKLFIHLREPLKVYSQNNRQFYYIKMSFNKYIKEDSAGLFLVNYYEDDVKNAKSLYATRLSPNKAKFMFPCFDNPRFEAVFKFKVYILSDHPGMQYTNSSLVIAEEMKRVSSKDYTIIEYIPSPQVALHQVGFHYSQFTNKSVKGKNNDTLVIWAPGDKLYCYDYIQRFGISMIDLMHQYEGTKRPITAGSINLVAVPTNLQGYEIGSWNLLTNGANRIANLVYITSIQQIEVMRFELAQQLCRIWLGNPGEENRTRWKEEWLKEGMATYYASYFLAQKTSERYPKRLQPLGIYGLSMKHKAMEIDWPRSTPALEVFNDTLAIEIPARYKDLVTMKTGTLFWMLENWIGPEKFHNALIKYINSRRGKYISLQDFLTTLDHETVECSYQLFNGSTSSRILRSWFSQAGYPVINVRVLRDRTPNVVQLNQRRFSFRNEHHRELEYLIPISYTIQNSKNCFNCYRPRFVIGSQPYTFIENLDGGWIILNSNGSGYYRVNYDSDSWLLIAKTLVEDHQSIEEMTRSQIVNDVFALYVSGNLDMSIAMQVLAYLDKETSLAVWDSVISGYELLKLEGAACNMTKYIYWEWSDFMKQKIPTIYNGLIKNLDQQTDVRLLRNKVITFAAELEYEQCLTQLRRMYQETRRDPKRILDPDARRGCYYVLLNDTNSHLRFKKLSGIEEEDRNSVENRIRENNRFLIRIPYGEPRPLPIISSESPLSSTTPEPVPDKIVMPDASTSNLSSIIFVVASILILRLF
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Feature
chain Aminopeptidase
Uniprot
H9JEW6
I3VR84
A0A2H1VVU9
A0A2W1BTL9
B8PYI4
A0A212EJS3
+ More
A0A194QCV3 G3ESV5 A0A3S2P7S7 A0A194QKB0 G3ESV6 E8Z7I6 A0A3G1QXU4 A0A194QD77 A0A194QK74 A0A2W1BPB2 B4PQ45 A0A2H1VVU0 I3VR76 B3P5I7 A0A0C9QRU9 B3MTC9 A0A0C9R4H9 B4QZJ3 Q29BX5 A0A2A4IXL1 K7J387 B3P5I6 B3P5I4 A0A232F800 B4GPI5 E9HEX4 A0A0N8AFY9 A0A0P5BZI3 A0A0P5BZD5 B4PQ42 A0A0R1E4S0 A0A0P5ASH2 A0A0P5E914 B4QZJ5 A0A0N8AGV7 A0A0P5VRM7 A0A1W4U620 A0A0N8CJQ2 A0A3B0JKK2 B3MTC8 A0A0P5GTI5 A0A0P6D560 B4HZ87 A0A0P5I561 A0A0P5IYG0 A0A1W4UH69 A0A0P5G1W8 A0A0P5FVD5 A0A0N8CNU5 A0A0P5JKV4 A0A0P5HXC8 A0A0K2UXX5 A0A0P5HP58 A0A0P5JLT0 A0A0P5IZL1 A0A195BIR1 A0A0P5LVC9 A0A2H8U053 A0A1W4XT18 B3NW11 A0A0P5N725 A0A2H8TQ38 Q9W2S7 A0A2J7RAG9 B4MBV1 E9HM93 Q6IDH1 D6WB37 B4IDS8 A0A0P5I5E6 Q0KI00 A0A1W4UH28 A0A146LYB5 A4D0J3 Q7QC91 A0A1L8E232 A0A1L8DPB4 B4JYJ5 A0A0K2UYY6 B4HZ86 A0A2J7RAH0 B4NFN7 A0A0Q9WV74 Q9NH67 A0A182HSV0 A0A0P5N2Y6 B4PQ46 A0A1B0GNF4 B3MTC6 A0A0P6A8B0 A0A182YDX4 B4QZJ4
A0A194QCV3 G3ESV5 A0A3S2P7S7 A0A194QKB0 G3ESV6 E8Z7I6 A0A3G1QXU4 A0A194QD77 A0A194QK74 A0A2W1BPB2 B4PQ45 A0A2H1VVU0 I3VR76 B3P5I7 A0A0C9QRU9 B3MTC9 A0A0C9R4H9 B4QZJ3 Q29BX5 A0A2A4IXL1 K7J387 B3P5I6 B3P5I4 A0A232F800 B4GPI5 E9HEX4 A0A0N8AFY9 A0A0P5BZI3 A0A0P5BZD5 B4PQ42 A0A0R1E4S0 A0A0P5ASH2 A0A0P5E914 B4QZJ5 A0A0N8AGV7 A0A0P5VRM7 A0A1W4U620 A0A0N8CJQ2 A0A3B0JKK2 B3MTC8 A0A0P5GTI5 A0A0P6D560 B4HZ87 A0A0P5I561 A0A0P5IYG0 A0A1W4UH69 A0A0P5G1W8 A0A0P5FVD5 A0A0N8CNU5 A0A0P5JKV4 A0A0P5HXC8 A0A0K2UXX5 A0A0P5HP58 A0A0P5JLT0 A0A0P5IZL1 A0A195BIR1 A0A0P5LVC9 A0A2H8U053 A0A1W4XT18 B3NW11 A0A0P5N725 A0A2H8TQ38 Q9W2S7 A0A2J7RAG9 B4MBV1 E9HM93 Q6IDH1 D6WB37 B4IDS8 A0A0P5I5E6 Q0KI00 A0A1W4UH28 A0A146LYB5 A4D0J3 Q7QC91 A0A1L8E232 A0A1L8DPB4 B4JYJ5 A0A0K2UYY6 B4HZ86 A0A2J7RAH0 B4NFN7 A0A0Q9WV74 Q9NH67 A0A182HSV0 A0A0P5N2Y6 B4PQ46 A0A1B0GNF4 B3MTC6 A0A0P6A8B0 A0A182YDX4 B4QZJ4
EC Number
3.4.11.-
Pubmed
EMBL
BABH01023392
BABH01023393
JQ061155
AFK85029.1
ODYU01004747
SOQ44950.1
+ More
KZ149969 PZC76120.1 EU328182 ACA35024.1 AGBW02014405 OWR41737.1 KQ459185 KPJ03373.1 JF339039 AEO12692.1 RSAL01000228 RVE43924.1 KQ461198 KPJ05972.1 AEO12693.1 FJ647796 ACV04931.1 KY949259 AWT23001.1 KPJ03374.1 KPJ05973.1 PZC76121.1 CM000160 EDW98314.1 SOQ44949.1 JQ061147 AFK85021.1 CH954182 EDV53237.1 GBYB01006459 JAG76226.1 CH902623 EDV30519.1 GBYB01002810 GBYB01007837 JAG72577.1 JAG77604.1 CM000364 EDX14831.1 CM000070 EAL26872.1 NWSH01005085 PCG64451.1 EDV53236.1 EDV53234.1 NNAY01000776 OXU26579.1 CH479186 EDW39068.1 GL732632 EFX69710.1 GDIP01151225 JAJ72177.1 GDIP01177791 JAJ45611.1 GDIP01177790 JAJ45612.1 EDW98311.1 KRK04205.1 GDIP01199474 JAJ23928.1 GDIP01149983 JAJ73419.1 EDX14833.1 GDIP01148681 JAJ74721.1 GDIP01096498 JAM07217.1 GDIP01132032 GDIP01125031 GDIP01111388 GDIP01007125 JAL78683.1 OUUW01000005 SPP81333.1 EDV30518.1 GDIQ01243285 GDIQ01241931 GDIQ01240487 GDIQ01222836 JAK08440.1 GDIQ01084835 JAN09902.1 CH480819 EDW53344.1 GDIQ01220101 JAK31624.1 GDIQ01208096 GDIQ01208095 GDIQ01208094 JAK43629.1 GDIQ01252386 GDIQ01252385 GDIQ01252384 JAJ99339.1 GDIQ01250982 GDIQ01250981 GDIQ01250980 GDIQ01080284 GDIQ01078385 JAK00743.1 GDIP01113487 JAL90227.1 GDIQ01218945 GDIQ01198349 JAK53376.1 GDIQ01244487 JAK07238.1 HACA01025559 CDW42920.1 GDIQ01249429 GDIQ01249428 GDIQ01247833 GDIQ01247832 GDIQ01241932 GDIQ01238793 GDIQ01234140 GDIQ01222837 GDIQ01221338 GDIQ01221337 GDIQ01204283 GDIQ01202201 GDIQ01202200 GDIQ01199651 GDIQ01195148 GDIQ01189688 GDIQ01189687 GDIQ01189686 GDIQ01188391 GDIQ01188390 GDIQ01188389 GDIQ01185244 GDIQ01181296 GDIQ01179943 GDIQ01162212 GDIQ01160810 GDIQ01159210 GDIQ01154856 GDIQ01146863 GDIQ01145273 GDIQ01143865 GDIQ01142315 GDIQ01140818 GDIQ01130136 GDIQ01128079 GDIQ01115734 GDIQ01113282 GDIQ01113281 GDIQ01111828 GDIQ01098582 GDIQ01096981 GDIQ01084834 GDIQ01084833 GDIQ01083333 GDIQ01081886 GDIQ01066996 GDIQ01066995 GDIQ01065798 GDIQ01064345 GDIQ01061046 GDIQ01051348 GDIQ01051347 GDIQ01048998 GDIQ01046700 GDIQ01035550 JAK03892.1 GDIQ01196896 JAK54829.1 GDIQ01207234 GDIQ01207233 GDIQ01207232 JAK44492.1 KQ976467 KYM84198.1 GDIQ01164902 GDIQ01021077 JAK86823.1 GFXV01008118 MBW19923.1 CH954180 EDV46144.1 GDIQ01168300 JAK83425.1 GFXV01004502 MBW16307.1 AE014298 AAF46612.1 NEVH01006570 PNF37826.1 CH940656 EDW58572.1 GL732685 EFX67103.1 BT014634 AAT27258.1 KQ971311 EEZ99336.1 CH480830 EDW45736.1 GDIQ01217792 JAK33933.1 AE014297 BT133378 AAF56884.1 AFH36363.1 GDHC01006045 JAQ12584.1 AY836579 AAX39863.1 AAAB01008859 EAA08230.5 GFDF01001291 JAV12793.1 GFDF01005795 JAV08289.1 CH916377 EDV90757.1 HACA01025560 CDW42921.1 EDW53343.1 PNF37825.1 CH964251 EDW83104.2 KRF99732.1 AF231040 AAF34809.1 APCN01000366 GDIQ01148024 GDIQ01148023 JAL03703.1 EDW98315.1 AJVK01029404 EDV30516.1 GDIP01037896 JAM65819.1 EDX14832.1
KZ149969 PZC76120.1 EU328182 ACA35024.1 AGBW02014405 OWR41737.1 KQ459185 KPJ03373.1 JF339039 AEO12692.1 RSAL01000228 RVE43924.1 KQ461198 KPJ05972.1 AEO12693.1 FJ647796 ACV04931.1 KY949259 AWT23001.1 KPJ03374.1 KPJ05973.1 PZC76121.1 CM000160 EDW98314.1 SOQ44949.1 JQ061147 AFK85021.1 CH954182 EDV53237.1 GBYB01006459 JAG76226.1 CH902623 EDV30519.1 GBYB01002810 GBYB01007837 JAG72577.1 JAG77604.1 CM000364 EDX14831.1 CM000070 EAL26872.1 NWSH01005085 PCG64451.1 EDV53236.1 EDV53234.1 NNAY01000776 OXU26579.1 CH479186 EDW39068.1 GL732632 EFX69710.1 GDIP01151225 JAJ72177.1 GDIP01177791 JAJ45611.1 GDIP01177790 JAJ45612.1 EDW98311.1 KRK04205.1 GDIP01199474 JAJ23928.1 GDIP01149983 JAJ73419.1 EDX14833.1 GDIP01148681 JAJ74721.1 GDIP01096498 JAM07217.1 GDIP01132032 GDIP01125031 GDIP01111388 GDIP01007125 JAL78683.1 OUUW01000005 SPP81333.1 EDV30518.1 GDIQ01243285 GDIQ01241931 GDIQ01240487 GDIQ01222836 JAK08440.1 GDIQ01084835 JAN09902.1 CH480819 EDW53344.1 GDIQ01220101 JAK31624.1 GDIQ01208096 GDIQ01208095 GDIQ01208094 JAK43629.1 GDIQ01252386 GDIQ01252385 GDIQ01252384 JAJ99339.1 GDIQ01250982 GDIQ01250981 GDIQ01250980 GDIQ01080284 GDIQ01078385 JAK00743.1 GDIP01113487 JAL90227.1 GDIQ01218945 GDIQ01198349 JAK53376.1 GDIQ01244487 JAK07238.1 HACA01025559 CDW42920.1 GDIQ01249429 GDIQ01249428 GDIQ01247833 GDIQ01247832 GDIQ01241932 GDIQ01238793 GDIQ01234140 GDIQ01222837 GDIQ01221338 GDIQ01221337 GDIQ01204283 GDIQ01202201 GDIQ01202200 GDIQ01199651 GDIQ01195148 GDIQ01189688 GDIQ01189687 GDIQ01189686 GDIQ01188391 GDIQ01188390 GDIQ01188389 GDIQ01185244 GDIQ01181296 GDIQ01179943 GDIQ01162212 GDIQ01160810 GDIQ01159210 GDIQ01154856 GDIQ01146863 GDIQ01145273 GDIQ01143865 GDIQ01142315 GDIQ01140818 GDIQ01130136 GDIQ01128079 GDIQ01115734 GDIQ01113282 GDIQ01113281 GDIQ01111828 GDIQ01098582 GDIQ01096981 GDIQ01084834 GDIQ01084833 GDIQ01083333 GDIQ01081886 GDIQ01066996 GDIQ01066995 GDIQ01065798 GDIQ01064345 GDIQ01061046 GDIQ01051348 GDIQ01051347 GDIQ01048998 GDIQ01046700 GDIQ01035550 JAK03892.1 GDIQ01196896 JAK54829.1 GDIQ01207234 GDIQ01207233 GDIQ01207232 JAK44492.1 KQ976467 KYM84198.1 GDIQ01164902 GDIQ01021077 JAK86823.1 GFXV01008118 MBW19923.1 CH954180 EDV46144.1 GDIQ01168300 JAK83425.1 GFXV01004502 MBW16307.1 AE014298 AAF46612.1 NEVH01006570 PNF37826.1 CH940656 EDW58572.1 GL732685 EFX67103.1 BT014634 AAT27258.1 KQ971311 EEZ99336.1 CH480830 EDW45736.1 GDIQ01217792 JAK33933.1 AE014297 BT133378 AAF56884.1 AFH36363.1 GDHC01006045 JAQ12584.1 AY836579 AAX39863.1 AAAB01008859 EAA08230.5 GFDF01001291 JAV12793.1 GFDF01005795 JAV08289.1 CH916377 EDV90757.1 HACA01025560 CDW42921.1 EDW53343.1 PNF37825.1 CH964251 EDW83104.2 KRF99732.1 AF231040 AAF34809.1 APCN01000366 GDIQ01148024 GDIQ01148023 JAL03703.1 EDW98315.1 AJVK01029404 EDV30516.1 GDIP01037896 JAM65819.1 EDX14832.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000283053
UP000053240
UP000002282
+ More
UP000008711 UP000007801 UP000000304 UP000001819 UP000218220 UP000002358 UP000215335 UP000008744 UP000000305 UP000192221 UP000268350 UP000001292 UP000078540 UP000192223 UP000000803 UP000235965 UP000008792 UP000007266 UP000007062 UP000001070 UP000007798 UP000075840 UP000092462 UP000076408
UP000008711 UP000007801 UP000000304 UP000001819 UP000218220 UP000002358 UP000215335 UP000008744 UP000000305 UP000192221 UP000268350 UP000001292 UP000078540 UP000192223 UP000000803 UP000235965 UP000008792 UP000007266 UP000007062 UP000001070 UP000007798 UP000075840 UP000092462 UP000076408
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JEW6
I3VR84
A0A2H1VVU9
A0A2W1BTL9
B8PYI4
A0A212EJS3
+ More
A0A194QCV3 G3ESV5 A0A3S2P7S7 A0A194QKB0 G3ESV6 E8Z7I6 A0A3G1QXU4 A0A194QD77 A0A194QK74 A0A2W1BPB2 B4PQ45 A0A2H1VVU0 I3VR76 B3P5I7 A0A0C9QRU9 B3MTC9 A0A0C9R4H9 B4QZJ3 Q29BX5 A0A2A4IXL1 K7J387 B3P5I6 B3P5I4 A0A232F800 B4GPI5 E9HEX4 A0A0N8AFY9 A0A0P5BZI3 A0A0P5BZD5 B4PQ42 A0A0R1E4S0 A0A0P5ASH2 A0A0P5E914 B4QZJ5 A0A0N8AGV7 A0A0P5VRM7 A0A1W4U620 A0A0N8CJQ2 A0A3B0JKK2 B3MTC8 A0A0P5GTI5 A0A0P6D560 B4HZ87 A0A0P5I561 A0A0P5IYG0 A0A1W4UH69 A0A0P5G1W8 A0A0P5FVD5 A0A0N8CNU5 A0A0P5JKV4 A0A0P5HXC8 A0A0K2UXX5 A0A0P5HP58 A0A0P5JLT0 A0A0P5IZL1 A0A195BIR1 A0A0P5LVC9 A0A2H8U053 A0A1W4XT18 B3NW11 A0A0P5N725 A0A2H8TQ38 Q9W2S7 A0A2J7RAG9 B4MBV1 E9HM93 Q6IDH1 D6WB37 B4IDS8 A0A0P5I5E6 Q0KI00 A0A1W4UH28 A0A146LYB5 A4D0J3 Q7QC91 A0A1L8E232 A0A1L8DPB4 B4JYJ5 A0A0K2UYY6 B4HZ86 A0A2J7RAH0 B4NFN7 A0A0Q9WV74 Q9NH67 A0A182HSV0 A0A0P5N2Y6 B4PQ46 A0A1B0GNF4 B3MTC6 A0A0P6A8B0 A0A182YDX4 B4QZJ4
A0A194QCV3 G3ESV5 A0A3S2P7S7 A0A194QKB0 G3ESV6 E8Z7I6 A0A3G1QXU4 A0A194QD77 A0A194QK74 A0A2W1BPB2 B4PQ45 A0A2H1VVU0 I3VR76 B3P5I7 A0A0C9QRU9 B3MTC9 A0A0C9R4H9 B4QZJ3 Q29BX5 A0A2A4IXL1 K7J387 B3P5I6 B3P5I4 A0A232F800 B4GPI5 E9HEX4 A0A0N8AFY9 A0A0P5BZI3 A0A0P5BZD5 B4PQ42 A0A0R1E4S0 A0A0P5ASH2 A0A0P5E914 B4QZJ5 A0A0N8AGV7 A0A0P5VRM7 A0A1W4U620 A0A0N8CJQ2 A0A3B0JKK2 B3MTC8 A0A0P5GTI5 A0A0P6D560 B4HZ87 A0A0P5I561 A0A0P5IYG0 A0A1W4UH69 A0A0P5G1W8 A0A0P5FVD5 A0A0N8CNU5 A0A0P5JKV4 A0A0P5HXC8 A0A0K2UXX5 A0A0P5HP58 A0A0P5JLT0 A0A0P5IZL1 A0A195BIR1 A0A0P5LVC9 A0A2H8U053 A0A1W4XT18 B3NW11 A0A0P5N725 A0A2H8TQ38 Q9W2S7 A0A2J7RAG9 B4MBV1 E9HM93 Q6IDH1 D6WB37 B4IDS8 A0A0P5I5E6 Q0KI00 A0A1W4UH28 A0A146LYB5 A4D0J3 Q7QC91 A0A1L8E232 A0A1L8DPB4 B4JYJ5 A0A0K2UYY6 B4HZ86 A0A2J7RAH0 B4NFN7 A0A0Q9WV74 Q9NH67 A0A182HSV0 A0A0P5N2Y6 B4PQ46 A0A1B0GNF4 B3MTC6 A0A0P6A8B0 A0A182YDX4 B4QZJ4
PDB
4WZ9
E-value=2.04893e-34,
Score=368
Ontologies
GO
PANTHER
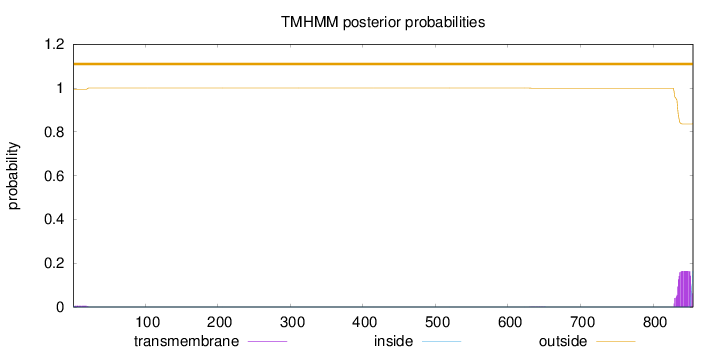
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.996782
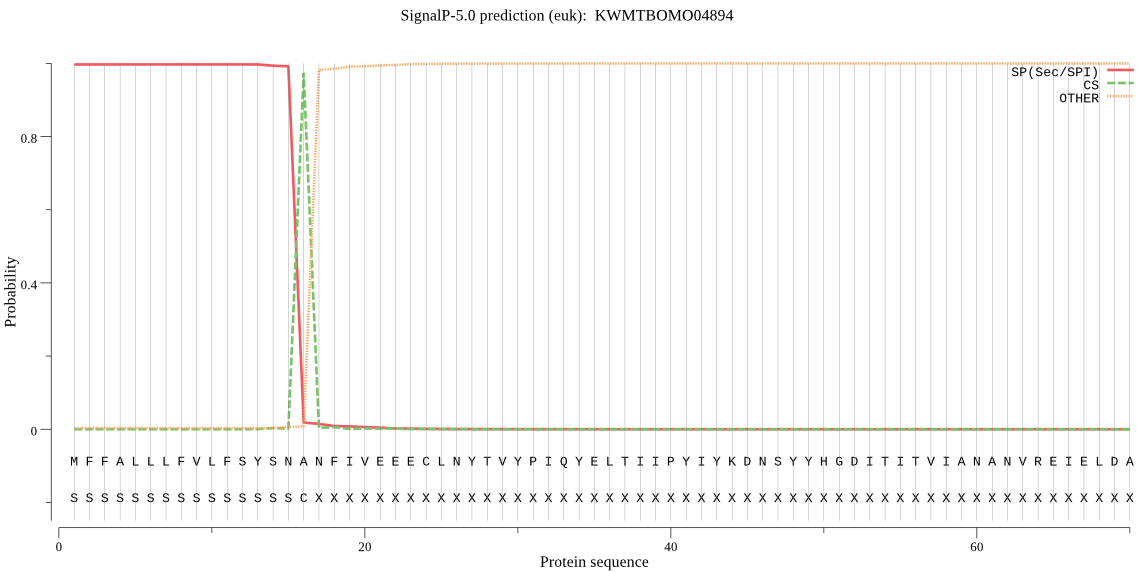
Length:
855
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.38924
Exp number, first 60 AAs:
0.09553
Total prob of N-in:
0.00507
outside
1 - 855
Population Genetic Test Statistics
Pi
191.859998
Theta
157.195429
Tajima's D
0.538066
CLR
0.457203
CSRT
0.522373881305935
Interpretation
Uncertain
