Gene
KWMTBOMO04891
Pre Gene Modal
BGIBMGA008061
Annotation
aminopeptidase_N-3_[Bombyx_mori]
Full name
Aminopeptidase
+ More
Membrane alanyl aminopeptidase
Membrane alanyl aminopeptidase
Alternative Name
Aminopeptidase N-like protein
CryIA(C) receptor
CryIA(C) receptor
Location in the cell
PlasmaMembrane Reliability : 3.402
Sequence
CDS
ATGTCCAAGCTTTTACTACTGTGTTTGGCGATTTTCGGCCTAACATCGGCTGACCACCCAGCATGGCACAGAGATTTCGAACCAATCCCCATAACTGACAATACAACGAGAAGCATTGAACAGGTTTATAGATTACCAGAGACAGTTAAACCATTGGAATACGATATTTATTTAGATTTGTACTTCGCTGAAAGAACAGACAGGCCCTTCAGTTATGATGGTAGAGAGACCATCATTATAGAGGCAGTAGCAGAAGGCGTGACTCAAATTGTTCTACATTCAAACGTGGACAGCATCAACGCTGTTTCTGTGCTAACGGAAGGTGGCGCACCATTCCCAGTTAATACAAGGGAGCCTTTCACACTGGAGCCTCAGTATCATTTCCTAAAAATTAATTTGAACGAACCCCTTGTTGTTGGACAGAAATACTCTTTGTATATTGAATATTCTAGCACAATGAACGAAGGCCCAATGAAGAGGGGTATTTGGAGGGGATGGTACATCGACAACGATGGTACAGAAAGGATCTACGCGACAACACATTTCCAGCCGTATAATGCGAGACAGGCATTCCCATGCTGGGACGAGCCACTGTTCAAGTCAGTCTTCAAACTCCACGTCTCGAAGCCAAAATCCTACACCGGCACGTTCTCAAACACTGGCGTCGAAACTTTTAGGTTTGAAGAAGCTGATCGCATTCGAAGCAATTTTCTGGCGACACCAAAAATGTCTTCGTACCTGGTTACGTTTCTTGTTAGCGAAACTTTCCAAGTATTGGCCGAGGACACTTCCTTCACTCCGCCTGTAAGGATCATTGGCCGGTCCAACACAGCAGGTCTTGCCGACCACGCATTGGAACTAGCAGTCCGTATGACCAGATACTATGATGAATATTTCGGGATTCCGTACTCAACGCTGCATCCAAATCTGCTCAATGATCATATTTCTTCCCCTGATTGGGCTTCGGCTGGCACAGAAAATTGGGGAATGGTCAGTTATAGGGAACTCTACCTCATCATTGATCCTCGTGAAACCGTCATGTCCGTGGAGCATTACGCTGCCACGTTGGTCTCCCACGAGCTCGCGCACAAATGGTTCGGTAACCTGATCACATGTTACTGGTGGAGCAACACTTGGATCAACGAAGGCTTTGCCAGTTACTTCGGGTACATTCCCACCTCTCAGATGTTTCCCGAGTATGAGCATCATGAGCACTTTAACGCCCGCTACGTTTCGTCATCGCTATCGTTCGACTCTGGCGTCAGCACCGTGCCTCTGAACCATGACGTAAACACACCAGCCCAAGTCACCGGCCACTTTGGAACCATCAGTTACTCTAAGGGCGCCGTATTCCTCAGGATGACTGCTGACATCATCACCGATGAGACTTTCCGAAAAGCTTGCCGTTACTTCTTGATTAATAACGCTTTCGAACCAACGGATCAATACGACTTATTGAAAGCTTTCAATCAAGCTATAATAGAAGACGGAAGTGTGTCTCAATATTCCAATTTTAACTTTGAAGAGTACTACCGCATTTGGGTCAACGAACCCGGATATCCGTTGCTGACGGTTACGGTTGACCATGGTGACGGAACTATTACTCTCAGTCAGGAACGATTCTTCTTGAGTGCATCTGCTACCCCGACAGACCATATATATCCAATTCCTATCACATATAGTAGTAAAAGCAATAATAACTTTGACAACGTCCGACCGGTCCATATAATGACCACTAAGACTGATGCTTTCAATGTTGGAGTTAAAGGAGAGTGGGTTATCTTCAACAATCTTCAACACGGACATTATAGAGTTACATATGATGATACAACATGGAATTTGATAGCAGATGCTTTGCTTAACGAAAGAGAGTCTATTCATCATCTAAATAGAGCTGAGATTGTGGATGACATCTTCGCCCTCATGAGGTCGGGCAGAATGACTTACAGTTTTGGTTTCAAAATCCTTCGCTTCTTGCGCAGTGAATCTAATTACCACGTGTGGGATGCAGCCATCACTGGCTACACTTGGTTGAGAAATAGACTCAGACATTTACCAGATCAAGCTACGTTTGACGCTTACATTCTGGAATTAATGGAAACAGTCATTAATACTTATGGCTTTGATGCCGCGGCTAATGAGCCTCCTACAACGAGTATGGCAAGACAAACTGTACTACAATTCGCTTGCAACCTTGGACACACAAAGTGCATCCAAGAATCGTACGACAAATTCAGAGAAATGAGAAGTGGTAAATGGGTTAGTCCTCAGATACGTCGTAACGTCTATATGACTGGCATGAGAGAGGGAGACAGTTCAGATTTCGAATACTTACTAAACCGTTTTCGACAATCGAACTTTGCTAACGATCAGCTAGAAATGCTCAGAGGATTGGGAGCTTCTAAAGATTCTCAATTATTGACTCGATACTTGCAGTTGACCTTGACTAGGGAGGTGAGATCGCACGATAAAGCTACTTCCTTCAACTACGCTCTGCTCGGTAACCAAGAGAACGCTAACACGGTTCTCCAGTTCGTGAAGAACAATATTGCTGCTATAAGAACGGCATACATTGAAGACGCTCCACCAACTCCAGTGCATACAGCTTTGTCTAATCTCGCTGCTTACCTTGATGAATCAGGTCTAGACGAATATGAAACTTGGCTACGTTCCACACAGACCAACATACCGCAGTACAATAGTGCGCTCTCAGCAATTAATTCGGCTCGGAGTAACATTGCTTGGGGCACTGCTAATGCCGAAATGTTATTAGCTGCTGCACGGGACAGTGCAACCGCAGTTGTGACGTCTGCTGCATTATTGGCTATAACTACACTGTTTGCTTTAATAATGTAA
Protein
MSKLLLLCLAIFGLTSADHPAWHRDFEPIPITDNTTRSIEQVYRLPETVKPLEYDIYLDLYFAERTDRPFSYDGRETIIIEAVAEGVTQIVLHSNVDSINAVSVLTEGGAPFPVNTREPFTLEPQYHFLKINLNEPLVVGQKYSLYIEYSSTMNEGPMKRGIWRGWYIDNDGTERIYATTHFQPYNARQAFPCWDEPLFKSVFKLHVSKPKSYTGTFSNTGVETFRFEEADRIRSNFLATPKMSSYLVTFLVSETFQVLAEDTSFTPPVRIIGRSNTAGLADHALELAVRMTRYYDEYFGIPYSTLHPNLLNDHISSPDWASAGTENWGMVSYRELYLIIDPRETVMSVEHYAATLVSHELAHKWFGNLITCYWWSNTWINEGFASYFGYIPTSQMFPEYEHHEHFNARYVSSSLSFDSGVSTVPLNHDVNTPAQVTGHFGTISYSKGAVFLRMTADIITDETFRKACRYFLINNAFEPTDQYDLLKAFNQAIIEDGSVSQYSNFNFEEYYRIWVNEPGYPLLTVTVDHGDGTITLSQERFFLSASATPTDHIYPIPITYSSKSNNNFDNVRPVHIMTTKTDAFNVGVKGEWVIFNNLQHGHYRVTYDDTTWNLIADALLNERESIHHLNRAEIVDDIFALMRSGRMTYSFGFKILRFLRSESNYHVWDAAITGYTWLRNRLRHLPDQATFDAYILELMETVINTYGFDAAANEPPTTSMARQTVLQFACNLGHTKCIQESYDKFREMRSGKWVSPQIRRNVYMTGMREGDSSDFEYLLNRFRQSNFANDQLEMLRGLGASKDSQLLTRYLQLTLTREVRSHDKATSFNYALLGNQENANTVLQFVKNNIAAIRTAYIEDAPPTPVHTALSNLAAYLDESGLDEYETWLRSTQTNIPQYNSALSAINSARSNIAWGTANAEMLLAAARDSATAVVTSAALLAITTLFALIM
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Keywords
Aminopeptidase
Cell membrane
Direct protein sequencing
Glycoprotein
GPI-anchor
Hydrolase
Lipoprotein
Membrane
Metal-binding
Metalloprotease
Protease
Signal
Zinc
Feature
chain Aminopeptidase
propeptide Activation peptide
propeptide Activation peptide
Uniprot
H9JEW4
I3VR74
A0A194QIU5
S4VAT7
A0A286LPZ8
A0A212EJS8
+ More
G0ZL62 B8PYI5 A0A2W1BM31 A0A2Z3YJ90 A0A2H1WZJ8 A0A2A4J7T7 A0A076VHN0 A0A194QR18 R9QYQ9 I3VR73 H9JEW5 A0A2A4IYF9 A0A194QEK1 S4VDN0 G9C5G3 Q7Z0W1 A0A2W1BR73 G3ESV7 Q6Q2I1 Q962B3 D3YJ04 D3YJ03 Q6PWP5 D2KHN2 A0A076YIK8 D2KHN1 O46156 A0A212EJS3 A0A076VDI4 G0ZL61 Q6J4J3 K9MBC9 Q0PDN2 Q963G0 A0A2W1BSE9 O77046 Q8MU79 Q7Z268 A4D0J6 A0A2A4J832 Q86QI6 Q8MUT5 Q8T5Q4 Q9NHZ8 G9C5G4 B2LS41 Q95WL1 B5A8X9 E8Z923 O16851 G9C7N4 A0A3S2P5X4 F2YWM1 D2KHN5 Q964F9 D2KHN4 H9JEW3 I3VR72 Q6UY55 Q4G6A2 Q962B4 Q6UY54 A0A2W1BR38 A0A212EJU6 A0A0L7L4F8 G3ESV3 A0A1B0RHN6 Q9U7N8 Q11001 E8Z5C9 D0UYB1 A0A1B0RHN3 O76803 E0A209 Q9U6W2 F6K6Z9 D0UYB2 A0A2H1WAU1 A0A1L5JK78 Q9NHZ9 B2LRS7 Q95WL0 Q6R3M5 Q6R3M6 B2LRS8 Q8WSZ2 A0A384RYH7 Q6T3W2 A4D0J3 H9JEW2 O76122 Q6T3W3
G0ZL62 B8PYI5 A0A2W1BM31 A0A2Z3YJ90 A0A2H1WZJ8 A0A2A4J7T7 A0A076VHN0 A0A194QR18 R9QYQ9 I3VR73 H9JEW5 A0A2A4IYF9 A0A194QEK1 S4VDN0 G9C5G3 Q7Z0W1 A0A2W1BR73 G3ESV7 Q6Q2I1 Q962B3 D3YJ04 D3YJ03 Q6PWP5 D2KHN2 A0A076YIK8 D2KHN1 O46156 A0A212EJS3 A0A076VDI4 G0ZL61 Q6J4J3 K9MBC9 Q0PDN2 Q963G0 A0A2W1BSE9 O77046 Q8MU79 Q7Z268 A4D0J6 A0A2A4J832 Q86QI6 Q8MUT5 Q8T5Q4 Q9NHZ8 G9C5G4 B2LS41 Q95WL1 B5A8X9 E8Z923 O16851 G9C7N4 A0A3S2P5X4 F2YWM1 D2KHN5 Q964F9 D2KHN4 H9JEW3 I3VR72 Q6UY55 Q4G6A2 Q962B4 Q6UY54 A0A2W1BR38 A0A212EJU6 A0A0L7L4F8 G3ESV3 A0A1B0RHN6 Q9U7N8 Q11001 E8Z5C9 D0UYB1 A0A1B0RHN3 O76803 E0A209 Q9U6W2 F6K6Z9 D0UYB2 A0A2H1WAU1 A0A1L5JK78 Q9NHZ9 B2LRS7 Q95WL0 Q6R3M5 Q6R3M6 B2LRS8 Q8WSZ2 A0A384RYH7 Q6T3W2 A4D0J3 H9JEW2 O76122 Q6T3W3
EC Number
3.4.11.-
Pubmed
EMBL
BABH01023391
BABH01023392
JQ061145
AFK85019.1
KQ459185
KPJ03376.1
+ More
KC776602 AGO57906.1 MF425655 ASU92547.1 AGBW02014405 OWR41736.1 JF303658 AEA29694.1 EU328183 ACA35025.1 KZ149969 PZC76119.1 KY949258 AWT23000.1 ODYU01012236 SOQ58437.1 NWSH01002538 PCG68031.1 KF900125 AIK27006.1 KQ461198 KPJ05976.1 JX101320 AGG36501.1 JQ061144 AFK85018.1 NWSH01005085 PCG64448.1 KPJ03375.1 KC776603 AGO57907.1 HQ853295 ADZ05467.1 AY279535 AY346383 AAP37951.1 AAQ24379.1 PZC76115.1 JF339039 AEO12694.1 AY564236 AAS75551.1 AY038608 AAK85539.1 GU479677 ADD39718.1 GU479676 ADD39717.1 AY573579 AAS82738.1 GU213037 ADA59489.1 KM034756 AIK67332.1 GU213036 ADA59488.1 AJ222699 CAA10950.1 OWR41737.1 KF900124 AIK27005.1 JF303657 AEA29693.1 AY601813 AAT45356.1 JQ088281 AFU51581.1 DQ872666 ABH07377.2 AF378666 AAK58066.1 PZC76117.1 AB013400 BAA33715.1 AF535165 AAN04899.1 AY279534 AAP37950.1 AY836582 AAX39866.1 PCG68029.1 AY181026 AAO23562.1 AF511038 AAM44056.1 AF498996 AAM18718.1 AF217249 AAF37559.1 HQ853296 ADZ05468.1 EU571948 ACB87202.2 AF317619 AAL26894.1 EU826126 ACF34998.2 FJ694793 ACV74256.1 AF020389 AAB70755.1 HQ901596 ADZ57273.1 RSAL01000014 RVE53191.1 JF509138 ADZ74247.1 GU213040 ADA59492.1 AF320764 AAK69605.1 GU213039 ADA59491.1 JQ061143 AFK85017.1 AY358034 AAQ57405.1 AY218845 AAP44967.1 AY038607 AAK85538.1 AY358035 AAQ57406.1 PZC76114.1 OWR41735.1 JTDY01003068 KOB70211.1 JF339038 AEO12690.1 KM360182 AKH49598.1 AF123313 AAF07223.1 X89081 AB007039 EU878375 ACJ64827.1 GQ927479 ACX85726.2 KM360190 AKH49606.1 AF084257 JQ061146 AAC33301.1 AFK85020.1 HM231316 ADL38968.1 AF173552 AAF08254.1 HM357836 AEA76302.1 GQ927480 ACX85727.2 ODYU01007373 SOQ50076.1 KY210882 APO15796.1 AF217248 AAF37558.1 EU568874 ACC68683.1 AF317620 AAL26895.1 AY515310 AAS00451.1 AY515309 AAS00450.1 EU568875 ACC68682.1 AF441377 AAL34109.1 KP053647 AKO62648.1 AY437833 AAR20814.1 AY836579 AAX39863.1 BABH01023388 BABH01023389 BABH01023390 AB007038 BAA32475.1 AY437832 AAR20813.1
KC776602 AGO57906.1 MF425655 ASU92547.1 AGBW02014405 OWR41736.1 JF303658 AEA29694.1 EU328183 ACA35025.1 KZ149969 PZC76119.1 KY949258 AWT23000.1 ODYU01012236 SOQ58437.1 NWSH01002538 PCG68031.1 KF900125 AIK27006.1 KQ461198 KPJ05976.1 JX101320 AGG36501.1 JQ061144 AFK85018.1 NWSH01005085 PCG64448.1 KPJ03375.1 KC776603 AGO57907.1 HQ853295 ADZ05467.1 AY279535 AY346383 AAP37951.1 AAQ24379.1 PZC76115.1 JF339039 AEO12694.1 AY564236 AAS75551.1 AY038608 AAK85539.1 GU479677 ADD39718.1 GU479676 ADD39717.1 AY573579 AAS82738.1 GU213037 ADA59489.1 KM034756 AIK67332.1 GU213036 ADA59488.1 AJ222699 CAA10950.1 OWR41737.1 KF900124 AIK27005.1 JF303657 AEA29693.1 AY601813 AAT45356.1 JQ088281 AFU51581.1 DQ872666 ABH07377.2 AF378666 AAK58066.1 PZC76117.1 AB013400 BAA33715.1 AF535165 AAN04899.1 AY279534 AAP37950.1 AY836582 AAX39866.1 PCG68029.1 AY181026 AAO23562.1 AF511038 AAM44056.1 AF498996 AAM18718.1 AF217249 AAF37559.1 HQ853296 ADZ05468.1 EU571948 ACB87202.2 AF317619 AAL26894.1 EU826126 ACF34998.2 FJ694793 ACV74256.1 AF020389 AAB70755.1 HQ901596 ADZ57273.1 RSAL01000014 RVE53191.1 JF509138 ADZ74247.1 GU213040 ADA59492.1 AF320764 AAK69605.1 GU213039 ADA59491.1 JQ061143 AFK85017.1 AY358034 AAQ57405.1 AY218845 AAP44967.1 AY038607 AAK85538.1 AY358035 AAQ57406.1 PZC76114.1 OWR41735.1 JTDY01003068 KOB70211.1 JF339038 AEO12690.1 KM360182 AKH49598.1 AF123313 AAF07223.1 X89081 AB007039 EU878375 ACJ64827.1 GQ927479 ACX85726.2 KM360190 AKH49606.1 AF084257 JQ061146 AAC33301.1 AFK85020.1 HM231316 ADL38968.1 AF173552 AAF08254.1 HM357836 AEA76302.1 GQ927480 ACX85727.2 ODYU01007373 SOQ50076.1 KY210882 APO15796.1 AF217248 AAF37558.1 EU568874 ACC68683.1 AF317620 AAL26895.1 AY515310 AAS00451.1 AY515309 AAS00450.1 EU568875 ACC68682.1 AF441377 AAL34109.1 KP053647 AKO62648.1 AY437833 AAR20814.1 AY836579 AAX39863.1 BABH01023388 BABH01023389 BABH01023390 AB007038 BAA32475.1 AY437832 AAR20813.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF48350
SSF48350
Gene 3D
CDD
ProteinModelPortal
H9JEW4
I3VR74
A0A194QIU5
S4VAT7
A0A286LPZ8
A0A212EJS8
+ More
G0ZL62 B8PYI5 A0A2W1BM31 A0A2Z3YJ90 A0A2H1WZJ8 A0A2A4J7T7 A0A076VHN0 A0A194QR18 R9QYQ9 I3VR73 H9JEW5 A0A2A4IYF9 A0A194QEK1 S4VDN0 G9C5G3 Q7Z0W1 A0A2W1BR73 G3ESV7 Q6Q2I1 Q962B3 D3YJ04 D3YJ03 Q6PWP5 D2KHN2 A0A076YIK8 D2KHN1 O46156 A0A212EJS3 A0A076VDI4 G0ZL61 Q6J4J3 K9MBC9 Q0PDN2 Q963G0 A0A2W1BSE9 O77046 Q8MU79 Q7Z268 A4D0J6 A0A2A4J832 Q86QI6 Q8MUT5 Q8T5Q4 Q9NHZ8 G9C5G4 B2LS41 Q95WL1 B5A8X9 E8Z923 O16851 G9C7N4 A0A3S2P5X4 F2YWM1 D2KHN5 Q964F9 D2KHN4 H9JEW3 I3VR72 Q6UY55 Q4G6A2 Q962B4 Q6UY54 A0A2W1BR38 A0A212EJU6 A0A0L7L4F8 G3ESV3 A0A1B0RHN6 Q9U7N8 Q11001 E8Z5C9 D0UYB1 A0A1B0RHN3 O76803 E0A209 Q9U6W2 F6K6Z9 D0UYB2 A0A2H1WAU1 A0A1L5JK78 Q9NHZ9 B2LRS7 Q95WL0 Q6R3M5 Q6R3M6 B2LRS8 Q8WSZ2 A0A384RYH7 Q6T3W2 A4D0J3 H9JEW2 O76122 Q6T3W3
G0ZL62 B8PYI5 A0A2W1BM31 A0A2Z3YJ90 A0A2H1WZJ8 A0A2A4J7T7 A0A076VHN0 A0A194QR18 R9QYQ9 I3VR73 H9JEW5 A0A2A4IYF9 A0A194QEK1 S4VDN0 G9C5G3 Q7Z0W1 A0A2W1BR73 G3ESV7 Q6Q2I1 Q962B3 D3YJ04 D3YJ03 Q6PWP5 D2KHN2 A0A076YIK8 D2KHN1 O46156 A0A212EJS3 A0A076VDI4 G0ZL61 Q6J4J3 K9MBC9 Q0PDN2 Q963G0 A0A2W1BSE9 O77046 Q8MU79 Q7Z268 A4D0J6 A0A2A4J832 Q86QI6 Q8MUT5 Q8T5Q4 Q9NHZ8 G9C5G4 B2LS41 Q95WL1 B5A8X9 E8Z923 O16851 G9C7N4 A0A3S2P5X4 F2YWM1 D2KHN5 Q964F9 D2KHN4 H9JEW3 I3VR72 Q6UY55 Q4G6A2 Q962B4 Q6UY54 A0A2W1BR38 A0A212EJU6 A0A0L7L4F8 G3ESV3 A0A1B0RHN6 Q9U7N8 Q11001 E8Z5C9 D0UYB1 A0A1B0RHN3 O76803 E0A209 Q9U6W2 F6K6Z9 D0UYB2 A0A2H1WAU1 A0A1L5JK78 Q9NHZ9 B2LRS7 Q95WL0 Q6R3M5 Q6R3M6 B2LRS8 Q8WSZ2 A0A384RYH7 Q6T3W2 A4D0J3 H9JEW2 O76122 Q6T3W3
PDB
4WZ9
E-value=5.05738e-101,
Score=943
Ontologies
GO
PANTHER
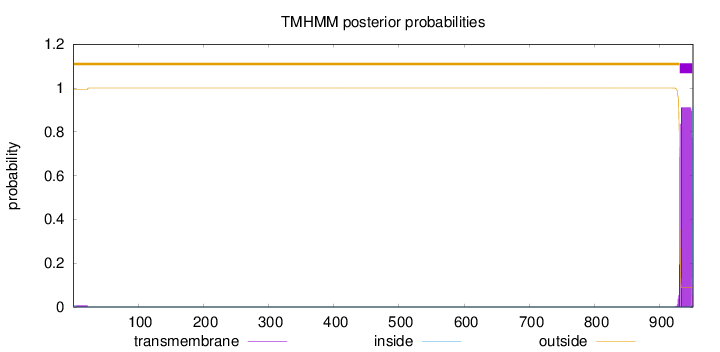
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 17,
Likelihood: 0.998326
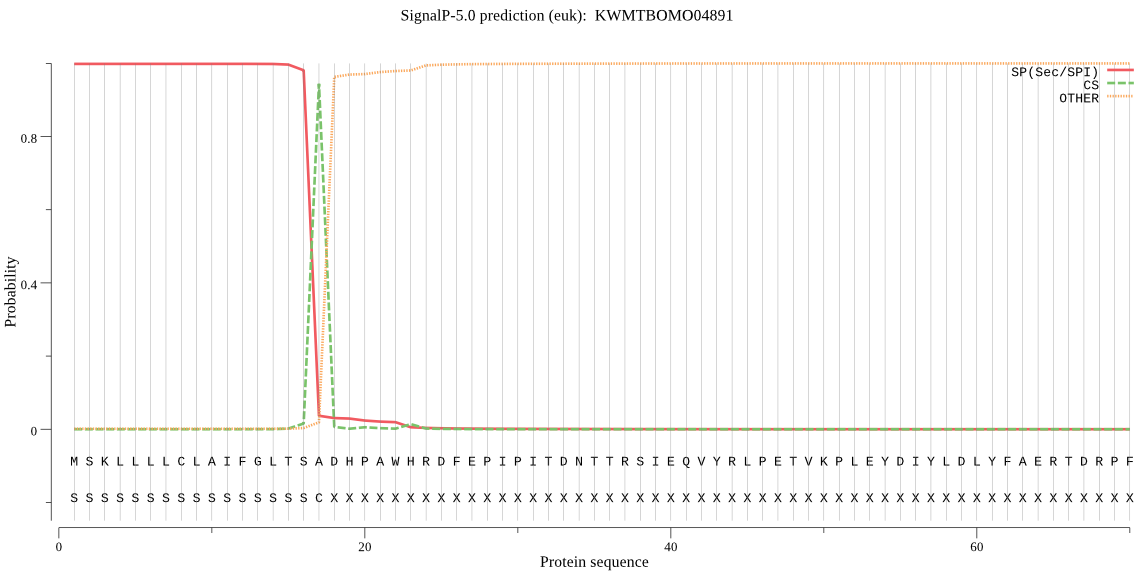
Length:
951
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.22569
Exp number, first 60 AAs:
0.14312
Total prob of N-in:
0.00789
outside
1 - 930
TMhelix
931 - 950
inside
951 - 951
Population Genetic Test Statistics
Pi
214.306356
Theta
209.309108
Tajima's D
-0.516143
CLR
0.238226
CSRT
0.239538023098845
Interpretation
Uncertain
