Gene
KWMTBOMO04886 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008059
Annotation
aminopeptidase_N-6_[Bombyx_mori]
Full name
Membrane alanyl aminopeptidase
+ More
Aminopeptidase
Aminopeptidase
Alternative Name
Aminopeptidase N-like protein
BTBP1
CryIA(C) receptor
BTBP1
CryIA(C) receptor
Location in the cell
PlasmaMembrane Reliability : 4.085
Sequence
CDS
ATGGCGAATTACAAGGTCATTATCTTCTTGGCGGCGTGCGTGTTAGCACAGGCTTTCCCCGATGAACCAATTTACAGGACAAACAACACAATATTTCTTGACGAGAAACTAGAAGGGGAAATCTTTGAGGATATTGAGGCTTTTGAAAATATTGATAGAAGTATTGCCGCTAGTACATACAGACTGCCCACAACCACCAGACCTCTTCATTACAACGTGCTTTGGGCTATTGATATCTCCAGGCTAACCTTCTCTGGCACCGTTGAAATCCAGCTGTACGCCACTCGGGCTAACGTAAGCGAGATAGTTATTCATGCCGACGACCTGGAAATCACATCCGTGATTCTCAGACAAGGAACCGTCACGACACCCTCCACTTACACCCTCCAGAAAGAGTTACAGTTCCTGAGACTGCGCCTTAATACGGGCACTCTCGTCTTCAATGCTGCTTCTCCTGTGATTTACACACTGACCATTGATTTCGCTGCTCGCTTACGTACCGACATGTATGGTATTTACCGGACCTGGTTCCGGAATAGCACTAATGACGTCACAAGATGGATGGCCTCTACACAGTTCCAAGCGACATCCGCTCGTTACGCTTTCCCTTGTTACGACGAGCCCAGTTTTAAAGCAACTTTTCATATTACGATCAGACGTCCGACCACCCACAGAAGTTGGTCGTGCACAAACATCAGAGAAACACGAGTTTCAACCGTAACGGGCTATCAAGATGACATTTACAACAGAACTCCATTAATGTCTACGTATTTGATTGCTCTTATTGTTGCTGAATACGAATCTTTGGAGCAGCGTCAAAACGGTGTCTTGAGCATCCACCCTAATTTAAAGATGACGCAGGCTTCAATTCCCGATTTTTCAGCTGGTGCTATGGAAAATTGGGGTCTGCTTACTTACAGAGAAGCATACTTAATGTACGATGAGAACCATACCAATGGCTACTTCAAACAGTTGATCGCTTACATCCTTTCTCACGAGATCGCCCATATGTGGTATGGAAACCTCGTTACTTGCGACTGGTGGGATGTTTTATGGCTGAACGAAGGTTTTGCTCGCTACTACCAATACTTCCTCACTGACTGGGTTGAAGATTACATGGGTCTTGGAACTCGATTCATTGTAGAGCAAATTCACACGTCCCTTTTGTCTGACTCGGCCAACAGCCCTCAGCCTTTGACCAACCCAGGTGTTGGCAGCCCCGCATCAGTGAGTGCCATGTTCTCGACCATTTCATACAACAAGGGTGCAGCTGTGATTAGAATGACTGAGCATTTCCTAGGCTTCGAAGTACATAGACAAGGCTTGAACAACTATCTGATTGAGAGATCTTTTGACACTGCTTTACCTATCCATCTCTTCCAAACCCTTGAAGTCAGCGCTAGAGCAGCTGGAGCTTTATCAGCATACGGACCAGATTTTTCTTTTGTTGACTATTACAAGAGTTGGACTGAACAGAGCGGTCACCCAGTACTTAACGTTCAAGTTAACCACCAGACTGGGGATATGACTATCTACCAGCGTCGTTTCAACATCAACACTGGTTACTCGAATGTCAACACCAACTACATCGTGCCTATCACTTTTGCCACTGCCCGCAACCCTAACTTCGCCAACACAAAGCCTACTCATGTCCTCACCAAAGCTGTTACTGTTATCAACAGAGGATCTGTTGGAGATGAATGGGTTATTTTCAATAAACAGCAGACTGGTTTCTACAGAGTCAACTATGATGACTACACTTGGAATCTCATTGTTATTGCTCTACGCGGCCCTCAAAGAACTCAAATTCACGAATACAATAGAGCACAGATCGTTAACGACGTCTTTCAATTCGCTCGCTCTGGTCTCATGACCTACAACAGAGCCTTCAACATTCTTTCTTTCCTTGAGAATGAAACTGAATACGCTCCGTGGGTCGCAGCTATTACTGGATTCAACTGGATCAGGAATCGTCTTGTGGGAACCGCCCACCTCACCACTTTGAATAATCTGATCGCCCGTTGGTCTTCGAATCTGATGAACCAGCTCACCTACTCTCCAATTCCCAACGAGTCCTTCATGAGGTCTTACCTCAGATATCAGCTGGCTCCTCTACTTTGTAACATCAACGTGGCAGCCTGTCGTACTGCAGCTACTACTCAGTTCCAAGCTCTTCGTGTTAACGGACAAGAAGTACCAGTGGACAATCGCAACTGGGTTTATTGCAACGCTCTTCGTGACGGCACTGAAGCGGACTTCAACTTTTTGTACCAGAGATTCCAAAGCCACGATGTATACACTGAAAAGATTCAAATCCTCTGGGTCCTTGGCTGCACTCCTCATGCTAACTCATTGAACACTCTTCTCAACGCCATCGTCCAGGATAACATCATCATCCGTCCTCAAGACTACACTAATGCCTTCAACAACGCTGTCTCTGGAAACGAAGGCAACACCCAGATAGCTTTCCGGTTCATCCAGAACAATTTGGCTGCCGTCACAGCTGCGTTCCAAAGTGTCGCAACTCCACTATCATACGTCTCATCCAGATTAAGAACGGAAGCTGAAATTGTTTCATTCCAAAATTGGGCGACTCAGAACCAGGTAGCGTTGGGTGATGCTTACCAAGCTGTGTTCCGCGGAGCCGAGACTTCCCGTGAAAGCATTGCTTGGGCTTCCTTAGTTCAAAACGACATGAACAGCTACTTCGTCACCGGAGACACGGTCTATGAAGCCTCTACTGCTGCAAACGTGATCACATCACCATCCACTACAGTAACTCCACCCAACCTGGTGGAGCCTGCGACTCCAAGCCTTCCAGTGCCTGATGCAGCACCAGTCTCTACTTTCTTATCTGTAGCTGTTGTGGCTTTAGTCGCTGTGGTTAACTTGATTATGTAG
Protein
MANYKVIIFLAACVLAQAFPDEPIYRTNNTIFLDEKLEGEIFEDIEAFENIDRSIAASTYRLPTTTRPLHYNVLWAIDISRLTFSGTVEIQLYATRANVSEIVIHADDLEITSVILRQGTVTTPSTYTLQKELQFLRLRLNTGTLVFNAASPVIYTLTIDFAARLRTDMYGIYRTWFRNSTNDVTRWMASTQFQATSARYAFPCYDEPSFKATFHITIRRPTTHRSWSCTNIRETRVSTVTGYQDDIYNRTPLMSTYLIALIVAEYESLEQRQNGVLSIHPNLKMTQASIPDFSAGAMENWGLLTYREAYLMYDENHTNGYFKQLIAYILSHEIAHMWYGNLVTCDWWDVLWLNEGFARYYQYFLTDWVEDYMGLGTRFIVEQIHTSLLSDSANSPQPLTNPGVGSPASVSAMFSTISYNKGAAVIRMTEHFLGFEVHRQGLNNYLIERSFDTALPIHLFQTLEVSARAAGALSAYGPDFSFVDYYKSWTEQSGHPVLNVQVNHQTGDMTIYQRRFNINTGYSNVNTNYIVPITFATARNPNFANTKPTHVLTKAVTVINRGSVGDEWVIFNKQQTGFYRVNYDDYTWNLIVIALRGPQRTQIHEYNRAQIVNDVFQFARSGLMTYNRAFNILSFLENETEYAPWVAAITGFNWIRNRLVGTAHLTTLNNLIARWSSNLMNQLTYSPIPNESFMRSYLRYQLAPLLCNINVAACRTAATTQFQALRVNGQEVPVDNRNWVYCNALRDGTEADFNFLYQRFQSHDVYTEKIQILWVLGCTPHANSLNTLLNAIVQDNIIIRPQDYTNAFNNAVSGNEGNTQIAFRFIQNNLAAVTAAFQSVATPLSYVSSRLRTEAEIVSFQNWATQNQVALGDAYQAVFRGAETSRESIAWASLVQNDMNSYFVTGDTVYEASTAANVITSPSTTVTPPNLVEPATPSLPVPDAAPVSTFLSVAVVALVAVVNLIM
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Keywords
Aminopeptidase
Cell membrane
Direct protein sequencing
Glycoprotein
GPI-anchor
Hydrolase
Lipoprotein
Membrane
Metal-binding
Metalloprotease
Protease
Signal
Zinc
Feature
propeptide Activation peptide
chain Membrane alanyl aminopeptidase
chain Membrane alanyl aminopeptidase
Uniprot
I3VR77
Q8T7M0
H9JEW2
Q8T3I0
A0A194QKB6
G3ESW0
+ More
G3ESV9 Q9GT75 E8Z9T8 E8Z5S9 A5JJU1 A7YAH7 G3ESV2 K9MBJ8 R4HHS0 E0A211 Q9NHZ7 M4M1W4 A0A0L7LCB7 Q9XYV2 Q11000 A4D0J5 G9C5G2 A0A3S2M847 H8YHV9 A0A096XIT2 Q7Z266 Q7Z267 A0A2W1BU00 Q8MU78 Q95UF9 A0A024AX72 A0A2Z3YJS3 Q6Y0Y6 A0A286LPZ6 Q9U8B0 Q8ITD3 O17485 O17484 A0A212EJT4 A0A2Z3YKX3 Q5UVJ2 Q4G6A3 A0A2H1V7F0 A1E3B6 A0A194QIU5 O16851 D2KHN5 D2KHN4 A0A212EJU6 Q19TV9 V5NQ96 V5NPR9 O76803 Q9NHZ9 Q962B4 Q4G6A5 F6K6Z9 A0A2W1BR38 Q9U6W2 B2LRS7 Q8WSZ2 Q95WL0 B2LRS8 A0A1L5JK78 A0A1B0RHN3 Q6UY54 Q6UY55 A0A1B0RHN6 Q11001 A0A384RYH7 Q9U7N8 E0A209 G3ESV3 Q6R3M5 Q6R3M6 D0UYB1 E8Z5C9 A4D0J3 Q6T3W2 O76122 D0UYB2 Q6T3W3 A0A3S2TRJ1 Q2L9Z0 A0A2H1WAG0 Q8ITD4 W8AMJ2 A0A3G1QXU4 A0A0K8U3X8 A0A0K8WLP7 A0A0A1XS95 A0A034WY81 A0A194QK74
G3ESV9 Q9GT75 E8Z9T8 E8Z5S9 A5JJU1 A7YAH7 G3ESV2 K9MBJ8 R4HHS0 E0A211 Q9NHZ7 M4M1W4 A0A0L7LCB7 Q9XYV2 Q11000 A4D0J5 G9C5G2 A0A3S2M847 H8YHV9 A0A096XIT2 Q7Z266 Q7Z267 A0A2W1BU00 Q8MU78 Q95UF9 A0A024AX72 A0A2Z3YJS3 Q6Y0Y6 A0A286LPZ6 Q9U8B0 Q8ITD3 O17485 O17484 A0A212EJT4 A0A2Z3YKX3 Q5UVJ2 Q4G6A3 A0A2H1V7F0 A1E3B6 A0A194QIU5 O16851 D2KHN5 D2KHN4 A0A212EJU6 Q19TV9 V5NQ96 V5NPR9 O76803 Q9NHZ9 Q962B4 Q4G6A5 F6K6Z9 A0A2W1BR38 Q9U6W2 B2LRS7 Q8WSZ2 Q95WL0 B2LRS8 A0A1L5JK78 A0A1B0RHN3 Q6UY54 Q6UY55 A0A1B0RHN6 Q11001 A0A384RYH7 Q9U7N8 E0A209 G3ESV3 Q6R3M5 Q6R3M6 D0UYB1 E8Z5C9 A4D0J3 Q6T3W2 O76122 D0UYB2 Q6T3W3 A0A3S2TRJ1 Q2L9Z0 A0A2H1WAG0 Q8ITD4 W8AMJ2 A0A3G1QXU4 A0A0K8U3X8 A0A0K8WLP7 A0A0A1XS95 A0A034WY81 A0A194QK74
EC Number
3.4.11.-
Pubmed
EMBL
JQ061148
AFK85022.1
AF352574
AAL83943.1
BABH01023388
BABH01023389
+ More
BABH01023390 AY095259 AAM13691.1 KQ461198 KPJ05977.1 JF339040 AEO12697.1 AEO12696.1 AF276241 AAF99701.1 FJ896130 ADA57169.1 FJ492806 ACT35083.1 EF538427 ABQ51393.1 EU137839 ABV01346.1 JF339038 AEO12689.1 JQ088280 AFU51580.1 HQ636624 AEL22855.1 HM231318 ADL38970.1 AF217250 AAF37560.1 JQ747495 AGG36452.1 JTDY01001695 KOB73138.1 AF126442 AAD31183.1 U35096 AY836581 AAX39865.1 HQ853294 ADZ05466.1 RSAL01000014 RVE53193.1 JF519739 AEB96253.1 KF689688 AHW57924.1 AY279537 AAP37953.1 AY279536 AAP37952.1 KZ149969 PZC76116.1 AF535166 AAN04900.1 AY052651 AAL14117.2 KJ143755 AHZ08543.1 KY949256 AWT22998.1 AY193789 AAP33526.1 MF425653 ASU92545.1 AF109692 AAF01259.2 AF521660 AAN75694.1 AF034484 AAC36147.1 AF034483 AAC36148.1 AGBW02014405 OWR41734.1 KY949255 AWT22997.1 AY603414 AAT99437.1 AY218844 AAP44966.1 ODYU01000825 SOQ36184.1 EF103944 ABL01483.1 KQ459185 KPJ03376.1 AF020389 AAB70755.1 GU213040 ADA59492.1 GU213039 ADA59491.1 OWR41735.1 DQ444715 ABE02186.1 KF537664 AHA90591.1 KF537663 AHA90590.1 AF084257 JQ061146 AAC33301.1 AFK85020.1 AF217248 AAF37558.1 AY038607 AAK85538.1 AY218842 AAP44964.1 HM357836 AEA76302.1 PZC76114.1 AF173552 AAF08254.1 EU568874 ACC68683.1 AF441377 AAL34109.1 AF317620 AAL26895.1 EU568875 ACC68682.1 KY210882 APO15796.1 KM360190 AKH49606.1 AY358035 AAQ57406.1 AY358034 AAQ57405.1 KM360182 AKH49598.1 X89081 AB007039 KP053647 AKO62648.1 AF123313 AAF07223.1 HM231316 ADL38968.1 AEO12690.1 AY515310 AAS00451.1 AY515309 AAS00450.1 GQ927479 ACX85726.2 EU878375 ACJ64827.1 AY836579 AAX39863.1 AY437833 AAR20814.1 AB007038 BAA32475.1 GQ927480 ACX85727.2 AY437832 AAR20813.1 RVE53192.1 DQ342305 ABC69855.3 ODYU01007373 SOQ50075.1 AF521659 AAN75693.1 GAMC01016580 JAB89975.1 KY949259 AWT23001.1 GDHF01031289 JAI21025.1 GDHF01000302 JAI52012.1 GBXI01000527 JAD13765.1 GAKP01000229 JAC58723.1 KPJ05973.1
BABH01023390 AY095259 AAM13691.1 KQ461198 KPJ05977.1 JF339040 AEO12697.1 AEO12696.1 AF276241 AAF99701.1 FJ896130 ADA57169.1 FJ492806 ACT35083.1 EF538427 ABQ51393.1 EU137839 ABV01346.1 JF339038 AEO12689.1 JQ088280 AFU51580.1 HQ636624 AEL22855.1 HM231318 ADL38970.1 AF217250 AAF37560.1 JQ747495 AGG36452.1 JTDY01001695 KOB73138.1 AF126442 AAD31183.1 U35096 AY836581 AAX39865.1 HQ853294 ADZ05466.1 RSAL01000014 RVE53193.1 JF519739 AEB96253.1 KF689688 AHW57924.1 AY279537 AAP37953.1 AY279536 AAP37952.1 KZ149969 PZC76116.1 AF535166 AAN04900.1 AY052651 AAL14117.2 KJ143755 AHZ08543.1 KY949256 AWT22998.1 AY193789 AAP33526.1 MF425653 ASU92545.1 AF109692 AAF01259.2 AF521660 AAN75694.1 AF034484 AAC36147.1 AF034483 AAC36148.1 AGBW02014405 OWR41734.1 KY949255 AWT22997.1 AY603414 AAT99437.1 AY218844 AAP44966.1 ODYU01000825 SOQ36184.1 EF103944 ABL01483.1 KQ459185 KPJ03376.1 AF020389 AAB70755.1 GU213040 ADA59492.1 GU213039 ADA59491.1 OWR41735.1 DQ444715 ABE02186.1 KF537664 AHA90591.1 KF537663 AHA90590.1 AF084257 JQ061146 AAC33301.1 AFK85020.1 AF217248 AAF37558.1 AY038607 AAK85538.1 AY218842 AAP44964.1 HM357836 AEA76302.1 PZC76114.1 AF173552 AAF08254.1 EU568874 ACC68683.1 AF441377 AAL34109.1 AF317620 AAL26895.1 EU568875 ACC68682.1 KY210882 APO15796.1 KM360190 AKH49606.1 AY358035 AAQ57406.1 AY358034 AAQ57405.1 KM360182 AKH49598.1 X89081 AB007039 KP053647 AKO62648.1 AF123313 AAF07223.1 HM231316 ADL38968.1 AEO12690.1 AY515310 AAS00451.1 AY515309 AAS00450.1 GQ927479 ACX85726.2 EU878375 ACJ64827.1 AY836579 AAX39863.1 AY437833 AAR20814.1 AB007038 BAA32475.1 GQ927480 ACX85727.2 AY437832 AAR20813.1 RVE53192.1 DQ342305 ABC69855.3 ODYU01007373 SOQ50075.1 AF521659 AAN75693.1 GAMC01016580 JAB89975.1 KY949259 AWT23001.1 GDHF01031289 JAI21025.1 GDHF01000302 JAI52012.1 GBXI01000527 JAD13765.1 GAKP01000229 JAC58723.1 KPJ05973.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
I3VR77
Q8T7M0
H9JEW2
Q8T3I0
A0A194QKB6
G3ESW0
+ More
G3ESV9 Q9GT75 E8Z9T8 E8Z5S9 A5JJU1 A7YAH7 G3ESV2 K9MBJ8 R4HHS0 E0A211 Q9NHZ7 M4M1W4 A0A0L7LCB7 Q9XYV2 Q11000 A4D0J5 G9C5G2 A0A3S2M847 H8YHV9 A0A096XIT2 Q7Z266 Q7Z267 A0A2W1BU00 Q8MU78 Q95UF9 A0A024AX72 A0A2Z3YJS3 Q6Y0Y6 A0A286LPZ6 Q9U8B0 Q8ITD3 O17485 O17484 A0A212EJT4 A0A2Z3YKX3 Q5UVJ2 Q4G6A3 A0A2H1V7F0 A1E3B6 A0A194QIU5 O16851 D2KHN5 D2KHN4 A0A212EJU6 Q19TV9 V5NQ96 V5NPR9 O76803 Q9NHZ9 Q962B4 Q4G6A5 F6K6Z9 A0A2W1BR38 Q9U6W2 B2LRS7 Q8WSZ2 Q95WL0 B2LRS8 A0A1L5JK78 A0A1B0RHN3 Q6UY54 Q6UY55 A0A1B0RHN6 Q11001 A0A384RYH7 Q9U7N8 E0A209 G3ESV3 Q6R3M5 Q6R3M6 D0UYB1 E8Z5C9 A4D0J3 Q6T3W2 O76122 D0UYB2 Q6T3W3 A0A3S2TRJ1 Q2L9Z0 A0A2H1WAG0 Q8ITD4 W8AMJ2 A0A3G1QXU4 A0A0K8U3X8 A0A0K8WLP7 A0A0A1XS95 A0A034WY81 A0A194QK74
G3ESV9 Q9GT75 E8Z9T8 E8Z5S9 A5JJU1 A7YAH7 G3ESV2 K9MBJ8 R4HHS0 E0A211 Q9NHZ7 M4M1W4 A0A0L7LCB7 Q9XYV2 Q11000 A4D0J5 G9C5G2 A0A3S2M847 H8YHV9 A0A096XIT2 Q7Z266 Q7Z267 A0A2W1BU00 Q8MU78 Q95UF9 A0A024AX72 A0A2Z3YJS3 Q6Y0Y6 A0A286LPZ6 Q9U8B0 Q8ITD3 O17485 O17484 A0A212EJT4 A0A2Z3YKX3 Q5UVJ2 Q4G6A3 A0A2H1V7F0 A1E3B6 A0A194QIU5 O16851 D2KHN5 D2KHN4 A0A212EJU6 Q19TV9 V5NQ96 V5NPR9 O76803 Q9NHZ9 Q962B4 Q4G6A5 F6K6Z9 A0A2W1BR38 Q9U6W2 B2LRS7 Q8WSZ2 Q95WL0 B2LRS8 A0A1L5JK78 A0A1B0RHN3 Q6UY54 Q6UY55 A0A1B0RHN6 Q11001 A0A384RYH7 Q9U7N8 E0A209 G3ESV3 Q6R3M5 Q6R3M6 D0UYB1 E8Z5C9 A4D0J3 Q6T3W2 O76122 D0UYB2 Q6T3W3 A0A3S2TRJ1 Q2L9Z0 A0A2H1WAG0 Q8ITD4 W8AMJ2 A0A3G1QXU4 A0A0K8U3X8 A0A0K8WLP7 A0A0A1XS95 A0A034WY81 A0A194QK74
PDB
4WZ9
E-value=1.40002e-103,
Score=965
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 18,
Likelihood: 0.987262
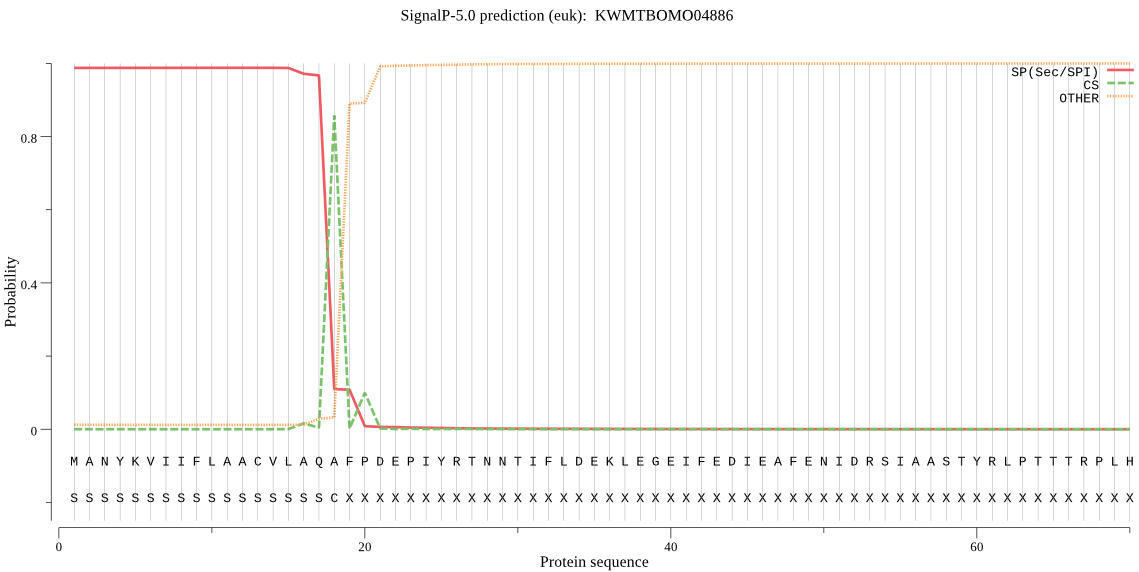
Length:
966
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
19.75723
Exp number, first 60 AAs:
0.28236
Total prob of N-in:
0.11928
outside
1 - 966
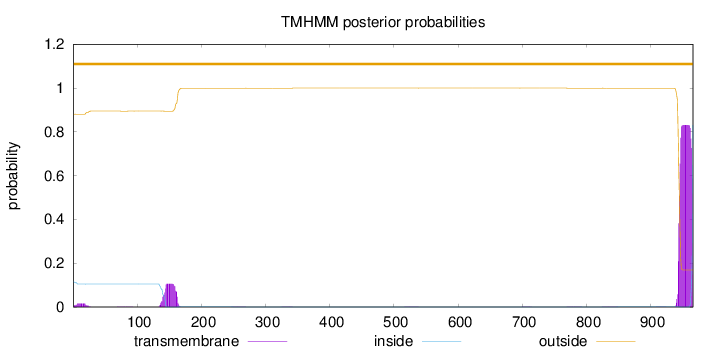
Population Genetic Test Statistics
Pi
226.691156
Theta
158.351975
Tajima's D
-1.431783
CLR
0.473208
CSRT
0.0694465276736163
Interpretation
Uncertain
