Gene
KWMTBOMO04885
Pre Gene Modal
BGIBMGA008058
Annotation
protease_m1_zinc_metalloprotease_[Danaus_plexippus]
Full name
Aminopeptidase
Location in the cell
PlasmaMembrane Reliability : 1.496
Sequence
CDS
ATGCAAACACATGAACCATTAAACGTAGTGGATGGCACTACGAAAACCGAAAGAGGAATCATTCTATCAAAGCCGTTAGTTCTATTTATATTCGTAATATTCGTACTGACTTTGGTGACGTCATCGCTTTTGGTGTACAATTTCGCTGCCTGCCCTAAAACTGATCAGCTGGCTAATGTCACAAAATACGAACTCTGTAGAGTTGCGGAAGCTAAACTGTCCCTTAATGAAACTAATAAAGATATACCTAAAGACAGGAATATAGAGGCTAACGATAAGAAATCAGCAACAGAAAGAAATATAGCTATAAGTAAGAATGAAATAGAGAAAGTCAAAGATGCAGAAGGTAAAGACGTCATGAACGATACAAATTTGGAGACACCTAAAGAATTTCGTTTACCTCAAAACGTGAAGCCGGAGAATTATTATCTAAACATTACTCCATTCTTCTTAGAGAATAATTTCACGTTTCAAGGCGAAGTTACTATATTGTTAACGGTTTTTAATGAGACTAATGAGATTATCTTCCATGGAGTCGATTTAGTGTTTAGAGATCTGAAGTTGCTGACAGAGAAAGATAGAAGAATAATAGACATCCTGAAAACAGAGGAAGATAAATTGAGGCAGTTTCATACAGTGACCGTTCGAGATAAATTAGAGGTTGGAATGCAGTATCTATTGGTAATAAGCTACTCAGGGGTCCTTAACGATGATCTCCATGGTTTTTATAGGAGCTCTTATGAAGACGGAGGCGTTAGAAAGTGGATAGCTGTGACGCAGTTCCAAGCTATGGACGCCCGGAGAGCGTTCCCTTGCTGGGACGAGCCGGCCATGAAGGCGCGGTTCACTATTACCATCGGAAGACCGGCCAATATGACTTCCTTGTCCAATATGAACAGTATTAAGCAGTCCCCACATGACAGTGTCGAAGGCTACATTAATGATTTTTACGAAGAATCATTGCCGATGTCGACGTATCTCGTCGCGTTTGCCGTCACAGATTTCAGTAGTAAATCGAAGGACACGTTCTCGGTGTGGGCGCGACATAAAGTCCTACCGGCCGCTTCTTTCGCCCTCGAAATCGGCTCGAAGATACTGAAGTTTCTAGAGAATTATTACGATATAAAATTTCCTTTACCGAAGATCGACATGATTGCTTTACCGGACTTCAAGAACGGTGCTATGGAGAACTGGGGACTCTTGACTTTCAGGGAAATAGCCATGCTTTACCAGCAAGGTGTGTCAGCTACGATGGACAAGGTGCACGTGGCCACCGTGGTCGCTCACGAGATCGCTCATCAGTGGTTCGGGAACCTCGTGACGCCTTCCTGGTGGCGAGACCTTTGGCTGAACGAGGGTTTCGCTACGTACGTCGAATACGTTGCTGTCGATGCTGTTGAGAAATCGTGGAATCTTACCGAGCTGTGTATTTTGGACCAAGTCCACAATGTATTCCAATTGGATGCGCTGAACTCTTCGCATCAGCTCTCCGTCGACGTCACGGCTTCCGAAGAGACCGACGCTATATTTGACAAAATATCATATGGAAAAGGTGCCGCTCTGCTCCGGATGCTGAACCACATTCTCACTGATGATATCTTCAACGCGGGCGTCACTAACTACCTGAAATCGAAGATGTATGGGAACGCGGAGCAGAAGGATCTCTGGGCTGCGCTGACTGCGGTCGCCACGCAAAGGAACCTCAAAATCGATGTTGCGGTCGTCATGGACTCGTGGACTTTGCAGACTGGATTCCCGGTGCTCACCGTCAATAGAGATTACGGAAACCAACAAATCAAATTCAGTCAGGAAAGGTTCGTACTGATCAATGAAACATCATCATCTCAACAAGCGCCCCTGTGGTGGATACCTGTGTCTTACACAACAGCTTCCGAGCAAGATTTCCGGTCGACCAGACCAAAGTTCTGGCTTGAGGGAGTGCAGAGTGTCGTCAAAGACTTCCCTGTCAAGAACGAGGACTGGTTAATTGTTAATATTCAAGAAACAGGTTTCTACCGCGTGAACTACGACACCCGTAACTGGCAACTCCTGACTGCGGTACTCAACGACAAGAATCGTTTTCAAGAAATCCACGTCATCAATCGAGCCCAGATAGTTGATGACGCGATGAACCTTGCTCTGACCGGTCGTTTGGACTACAAGACGGCCTTGGACGTGGTCAGCTACCTCGCCCATGAACGCAGCTACGTACCATGGAAGGCCGGTCTTTCTGCCCTTGGCTATATTGACACCATGCTGTCAAATGGAGAGCATTATGTGGAATACGGGATATACGTTGATCGGCTGCTACAAGGGGCCATCAGAGATGTCGGATGGGACATTCGAGAAGACGAGAGTGTTATAACGGCCCAGCACAGGGTGGATCTCTGGGCATCGGCGTGCCATTTCGAGAATTCAGACTGCATACAGAATGCTATTATATCATTCAAGAATTGGAGCACTTCTCCTGACCCTGACCGCTTTAATGATATACCGGTGGACGTGCGCGGGATAGTGTACTGCATGGCGCTGCGGCTGGGCGGCGTCAGCGAGTGGCAGTTCGCGCTGGACCGCCTCCCCGCCGCCGCGCCCGCCGAGCGACACCGCCTGCTCTCCGTGCTCGGCTGCACCAGGACCCCGCACCTACTGCGCAGGTACTTGGACATGTCATTGCGGAACGACAGTGGCATACGGAAGCAGGACGTGGTGCGGGTCTTCTCTGCTGTCTCCAACACTGCCATAGGACAGCCCATATCCTTCAGCTTCATCAGAGAGAACTGGCAGAATATCAGAACGTATTTCGGGTCAATATCAACTTTAAACCACATCGTGAAGGTCGTGACTCGCAGACTGAATCTACAACATGAATATGAAGAGCTGAAGAGGTTCGTCTCGGAGTCTTGTAGCGACCTGGGACGTCCAGTGCGGCAGGTCCTGGAGACAGTACACGCGAACGTTCAGTGGAGAAATAGAAATTATCAAACCATTGTAGATTGGCTGCAGGAAGTTAATAAAAAATACAAGGCTTAA
Protein
MQTHEPLNVVDGTTKTERGIILSKPLVLFIFVIFVLTLVTSSLLVYNFAACPKTDQLANVTKYELCRVAEAKLSLNETNKDIPKDRNIEANDKKSATERNIAISKNEIEKVKDAEGKDVMNDTNLETPKEFRLPQNVKPENYYLNITPFFLENNFTFQGEVTILLTVFNETNEIIFHGVDLVFRDLKLLTEKDRRIIDILKTEEDKLRQFHTVTVRDKLEVGMQYLLVISYSGVLNDDLHGFYRSSYEDGGVRKWIAVTQFQAMDARRAFPCWDEPAMKARFTITIGRPANMTSLSNMNSIKQSPHDSVEGYINDFYEESLPMSTYLVAFAVTDFSSKSKDTFSVWARHKVLPAASFALEIGSKILKFLENYYDIKFPLPKIDMIALPDFKNGAMENWGLLTFREIAMLYQQGVSATMDKVHVATVVAHEIAHQWFGNLVTPSWWRDLWLNEGFATYVEYVAVDAVEKSWNLTELCILDQVHNVFQLDALNSSHQLSVDVTASEETDAIFDKISYGKGAALLRMLNHILTDDIFNAGVTNYLKSKMYGNAEQKDLWAALTAVATQRNLKIDVAVVMDSWTLQTGFPVLTVNRDYGNQQIKFSQERFVLINETSSSQQAPLWWIPVSYTTASEQDFRSTRPKFWLEGVQSVVKDFPVKNEDWLIVNIQETGFYRVNYDTRNWQLLTAVLNDKNRFQEIHVINRAQIVDDAMNLALTGRLDYKTALDVVSYLAHERSYVPWKAGLSALGYIDTMLSNGEHYVEYGIYVDRLLQGAIRDVGWDIREDESVITAQHRVDLWASACHFENSDCIQNAIISFKNWSTSPDPDRFNDIPVDVRGIVYCMALRLGGVSEWQFALDRLPAAAPAERHRLLSVLGCTRTPHLLRRYLDMSLRNDSGIRKQDVVRVFSAVSNTAIGQPISFSFIRENWQNIRTYFGSISTLNHIVKVVTRRLNLQHEYEELKRFVSESCSDLGRPVRQVLETVHANVQWRNRNYQTIVDWLQEVNKKYKA
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Uniprot
A0A2H1V5Y2
A0A2W1BWK3
A0A2A4J2H3
A0A212EJQ4
A0A194QK79
A0A194QD38
+ More
A0A3S2PJF9 A0A2A4ITR7 E2BB79 K7INE5 E2ALT1 A0A1Q3FZ21 A0A1Q3FZ53 A0A084WR42 A0A182R8C8 A0A158P3F3 A0A182NCK3 A0A182GA80 A0A195F0B1 Q178P2 A0A182IV65 A0A026W0P7 A0A232EWK0 A0A3L8E4L7 A0A182YG74 A0A087ZUG8 A0A2A3EJS2 A0A182Q4G3 A0A182T1W9 A0A2J7RAG3 F5HLF5 A0A182F8E0 A0A0M9A6N6 A0A182PTT0 A0A0L7QWL2 A0A182KFB7 A0A182W2Z0 A0A182U9P3 A0A182VAQ1 A0A182WVF9 A0A182I938 W5JHD0 A0A195BIK5 A0A195DN33 A0A1S4FBS0 A0A1B0CGE3 A0A336MBJ6 A0A336M794 A0A336KRZ8 A0A336KP62 A0A1L8E2W2 A0A1L8E2E6 A0A067QSR5 A0A195CCY0 A0A151WZ01 A0A1B6CLC6 A0A0L0C6T3 A0A034V8K5 F4W4E5 A0A195DN37 E9JC17 A0A0K8UB09 E0VS71 A0A0A1WFD9 A0A0P4VSA7 A0A2R7VTS0 A0A069DXK6 A0A224X6F7 A0A154PMQ1 A0A1B6M3I9 A0A146LYB5 A0A0V0G738 A0A1B6GWM4 A0A182KXY3 T1P8B3 A0A1I8MFJ3 T1I7S8 A0A1I8P6R0 A0A1Y1LDQ6 A0A1Y1L923 A0A0R3NGA2 B5DWU2 A0A0M4F5M5 A0A1B6E6H9 B4G2I1 A0A3B0K3P3 D6WCY0 B3MT48 W8BB89 A0A1W4UIQ0 A0A1W4U5E1 A0A1W4UGV9 B4NBG4 B4LYI7 B4K8N1 A0A1J1I8E4 A0A195BIR1 Q7KRW4 A0A310SPN7 B4JTM7
A0A3S2PJF9 A0A2A4ITR7 E2BB79 K7INE5 E2ALT1 A0A1Q3FZ21 A0A1Q3FZ53 A0A084WR42 A0A182R8C8 A0A158P3F3 A0A182NCK3 A0A182GA80 A0A195F0B1 Q178P2 A0A182IV65 A0A026W0P7 A0A232EWK0 A0A3L8E4L7 A0A182YG74 A0A087ZUG8 A0A2A3EJS2 A0A182Q4G3 A0A182T1W9 A0A2J7RAG3 F5HLF5 A0A182F8E0 A0A0M9A6N6 A0A182PTT0 A0A0L7QWL2 A0A182KFB7 A0A182W2Z0 A0A182U9P3 A0A182VAQ1 A0A182WVF9 A0A182I938 W5JHD0 A0A195BIK5 A0A195DN33 A0A1S4FBS0 A0A1B0CGE3 A0A336MBJ6 A0A336M794 A0A336KRZ8 A0A336KP62 A0A1L8E2W2 A0A1L8E2E6 A0A067QSR5 A0A195CCY0 A0A151WZ01 A0A1B6CLC6 A0A0L0C6T3 A0A034V8K5 F4W4E5 A0A195DN37 E9JC17 A0A0K8UB09 E0VS71 A0A0A1WFD9 A0A0P4VSA7 A0A2R7VTS0 A0A069DXK6 A0A224X6F7 A0A154PMQ1 A0A1B6M3I9 A0A146LYB5 A0A0V0G738 A0A1B6GWM4 A0A182KXY3 T1P8B3 A0A1I8MFJ3 T1I7S8 A0A1I8P6R0 A0A1Y1LDQ6 A0A1Y1L923 A0A0R3NGA2 B5DWU2 A0A0M4F5M5 A0A1B6E6H9 B4G2I1 A0A3B0K3P3 D6WCY0 B3MT48 W8BB89 A0A1W4UIQ0 A0A1W4U5E1 A0A1W4UGV9 B4NBG4 B4LYI7 B4K8N1 A0A1J1I8E4 A0A195BIR1 Q7KRW4 A0A310SPN7 B4JTM7
EC Number
3.4.11.-
Pubmed
28756777
22118469
26354079
20798317
20075255
24438588
+ More
21347285 26483478 17510324 24508170 28648823 30249741 25244985 12364791 14747013 17210077 20920257 23761445 24845553 26108605 25348373 21719571 21282665 20566863 25830018 27129103 26334808 26823975 20966253 25315136 28004739 15632085 17994087 18362917 19820115 18057021 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
21347285 26483478 17510324 24508170 28648823 30249741 25244985 12364791 14747013 17210077 20920257 23761445 24845553 26108605 25348373 21719571 21282665 20566863 25830018 27129103 26334808 26823975 20966253 25315136 28004739 15632085 17994087 18362917 19820115 18057021 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
EMBL
ODYU01000825
SOQ36186.1
KZ149969
PZC76123.1
NWSH01003997
PCG65583.1
+ More
AGBW02014405 OWR41732.1 KQ461198 KPJ05978.1 KQ459185 KPJ03377.1 RSAL01000014 RVE53196.1 NWSH01007621 PCG62886.1 GL446946 EFN87052.1 GL440629 EFN65601.1 GFDL01002220 JAV32825.1 GFDL01002200 JAV32845.1 ATLV01025918 KE525402 KFB52686.1 ADTU01007781 ADTU01007782 ADTU01007783 JXUM01050786 JXUM01050787 JXUM01050788 JXUM01050789 KQ561662 KXJ77885.1 KQ981905 KYN33597.1 CH477360 EAT42699.1 KK107589 EZA48634.1 NNAY01001852 OXU22728.1 QOIP01000001 RLU27566.1 KZ288223 PBC31977.1 AXCN02001184 NEVH01006570 PNF37828.1 AAAB01008888 EGK97116.1 KQ435724 KOX78330.1 KQ414710 KOC62990.1 APCN01003169 ADMH02001252 ETN63471.1 KQ976467 KYM84197.1 KQ980713 KYN14242.1 AJWK01011047 AJWK01011048 AJWK01011049 UFQS01000655 UFQT01000655 SSX05824.1 SSX26183.1 SSX05825.1 SSX26184.1 SSX05823.1 SSX26182.1 SSX05822.1 SSX26181.1 GFDF01001172 JAV12912.1 GFDF01001171 JAV12913.1 KK853596 KDR06435.1 KQ977935 KYM98719.1 KQ982649 KYQ53113.1 GEDC01023029 GEDC01011996 JAS14269.1 JAS25302.1 JRES01000835 KNC27946.1 GAKP01020867 JAC38085.1 GL887532 EGI70840.1 KYN14241.1 GL771642 EFZ09701.1 GDHF01028490 GDHF01018031 GDHF01005941 JAI23824.1 JAI34283.1 JAI46373.1 DS235745 EEB16227.1 GBXI01017159 JAC97132.1 GDKW01001137 JAI55458.1 KK854013 PTY09335.1 GBGD01000333 JAC88556.1 GFTR01008371 JAW08055.1 KQ434984 KZC13132.1 GEBQ01009519 JAT30458.1 GDHC01006045 JAQ12584.1 GECL01002882 JAP03242.1 GECZ01002996 JAS66773.1 KA644849 AFP59478.1 ACPB03005577 GEZM01062043 JAV70095.1 GEZM01062044 JAV70094.1 CM000070 KRT00027.1 EDY67826.1 CP012526 ALC47212.1 GEDC01003748 JAS33550.1 CH479179 EDW24026.1 OUUW01000005 SPP80599.1 KQ971311 EEZ99360.2 CH902623 EDV30438.1 KPU72858.1 GAMC01012267 JAB94288.1 CH964232 EDW81128.2 CH940650 EDW68007.1 CH933806 EDW15450.1 CVRI01000044 CRK96557.1 KYM84198.1 AE014297 AAN14161.2 KQ760397 OAD60666.1 CH916374 EDV91456.1
AGBW02014405 OWR41732.1 KQ461198 KPJ05978.1 KQ459185 KPJ03377.1 RSAL01000014 RVE53196.1 NWSH01007621 PCG62886.1 GL446946 EFN87052.1 GL440629 EFN65601.1 GFDL01002220 JAV32825.1 GFDL01002200 JAV32845.1 ATLV01025918 KE525402 KFB52686.1 ADTU01007781 ADTU01007782 ADTU01007783 JXUM01050786 JXUM01050787 JXUM01050788 JXUM01050789 KQ561662 KXJ77885.1 KQ981905 KYN33597.1 CH477360 EAT42699.1 KK107589 EZA48634.1 NNAY01001852 OXU22728.1 QOIP01000001 RLU27566.1 KZ288223 PBC31977.1 AXCN02001184 NEVH01006570 PNF37828.1 AAAB01008888 EGK97116.1 KQ435724 KOX78330.1 KQ414710 KOC62990.1 APCN01003169 ADMH02001252 ETN63471.1 KQ976467 KYM84197.1 KQ980713 KYN14242.1 AJWK01011047 AJWK01011048 AJWK01011049 UFQS01000655 UFQT01000655 SSX05824.1 SSX26183.1 SSX05825.1 SSX26184.1 SSX05823.1 SSX26182.1 SSX05822.1 SSX26181.1 GFDF01001172 JAV12912.1 GFDF01001171 JAV12913.1 KK853596 KDR06435.1 KQ977935 KYM98719.1 KQ982649 KYQ53113.1 GEDC01023029 GEDC01011996 JAS14269.1 JAS25302.1 JRES01000835 KNC27946.1 GAKP01020867 JAC38085.1 GL887532 EGI70840.1 KYN14241.1 GL771642 EFZ09701.1 GDHF01028490 GDHF01018031 GDHF01005941 JAI23824.1 JAI34283.1 JAI46373.1 DS235745 EEB16227.1 GBXI01017159 JAC97132.1 GDKW01001137 JAI55458.1 KK854013 PTY09335.1 GBGD01000333 JAC88556.1 GFTR01008371 JAW08055.1 KQ434984 KZC13132.1 GEBQ01009519 JAT30458.1 GDHC01006045 JAQ12584.1 GECL01002882 JAP03242.1 GECZ01002996 JAS66773.1 KA644849 AFP59478.1 ACPB03005577 GEZM01062043 JAV70095.1 GEZM01062044 JAV70094.1 CM000070 KRT00027.1 EDY67826.1 CP012526 ALC47212.1 GEDC01003748 JAS33550.1 CH479179 EDW24026.1 OUUW01000005 SPP80599.1 KQ971311 EEZ99360.2 CH902623 EDV30438.1 KPU72858.1 GAMC01012267 JAB94288.1 CH964232 EDW81128.2 CH940650 EDW68007.1 CH933806 EDW15450.1 CVRI01000044 CRK96557.1 KYM84198.1 AE014297 AAN14161.2 KQ760397 OAD60666.1 CH916374 EDV91456.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
UP000008237
+ More
UP000002358 UP000000311 UP000030765 UP000075900 UP000005205 UP000075884 UP000069940 UP000249989 UP000078541 UP000008820 UP000075880 UP000053097 UP000215335 UP000279307 UP000076408 UP000005203 UP000242457 UP000075886 UP000075901 UP000235965 UP000007062 UP000069272 UP000053105 UP000075885 UP000053825 UP000075881 UP000075920 UP000075902 UP000075903 UP000076407 UP000075840 UP000000673 UP000078540 UP000078492 UP000092461 UP000027135 UP000078542 UP000075809 UP000037069 UP000007755 UP000009046 UP000076502 UP000075882 UP000095301 UP000015103 UP000095300 UP000001819 UP000092553 UP000008744 UP000268350 UP000007266 UP000007801 UP000192221 UP000007798 UP000008792 UP000009192 UP000183832 UP000000803 UP000001070
UP000002358 UP000000311 UP000030765 UP000075900 UP000005205 UP000075884 UP000069940 UP000249989 UP000078541 UP000008820 UP000075880 UP000053097 UP000215335 UP000279307 UP000076408 UP000005203 UP000242457 UP000075886 UP000075901 UP000235965 UP000007062 UP000069272 UP000053105 UP000075885 UP000053825 UP000075881 UP000075920 UP000075902 UP000075903 UP000076407 UP000075840 UP000000673 UP000078540 UP000078492 UP000092461 UP000027135 UP000078542 UP000075809 UP000037069 UP000007755 UP000009046 UP000076502 UP000075882 UP000095301 UP000015103 UP000095300 UP000001819 UP000092553 UP000008744 UP000268350 UP000007266 UP000007801 UP000192221 UP000007798 UP000008792 UP000009192 UP000183832 UP000000803 UP000001070
PRIDE
Interpro
SUPFAM
SSF50494
SSF50494
Gene 3D
ProteinModelPortal
A0A2H1V5Y2
A0A2W1BWK3
A0A2A4J2H3
A0A212EJQ4
A0A194QK79
A0A194QD38
+ More
A0A3S2PJF9 A0A2A4ITR7 E2BB79 K7INE5 E2ALT1 A0A1Q3FZ21 A0A1Q3FZ53 A0A084WR42 A0A182R8C8 A0A158P3F3 A0A182NCK3 A0A182GA80 A0A195F0B1 Q178P2 A0A182IV65 A0A026W0P7 A0A232EWK0 A0A3L8E4L7 A0A182YG74 A0A087ZUG8 A0A2A3EJS2 A0A182Q4G3 A0A182T1W9 A0A2J7RAG3 F5HLF5 A0A182F8E0 A0A0M9A6N6 A0A182PTT0 A0A0L7QWL2 A0A182KFB7 A0A182W2Z0 A0A182U9P3 A0A182VAQ1 A0A182WVF9 A0A182I938 W5JHD0 A0A195BIK5 A0A195DN33 A0A1S4FBS0 A0A1B0CGE3 A0A336MBJ6 A0A336M794 A0A336KRZ8 A0A336KP62 A0A1L8E2W2 A0A1L8E2E6 A0A067QSR5 A0A195CCY0 A0A151WZ01 A0A1B6CLC6 A0A0L0C6T3 A0A034V8K5 F4W4E5 A0A195DN37 E9JC17 A0A0K8UB09 E0VS71 A0A0A1WFD9 A0A0P4VSA7 A0A2R7VTS0 A0A069DXK6 A0A224X6F7 A0A154PMQ1 A0A1B6M3I9 A0A146LYB5 A0A0V0G738 A0A1B6GWM4 A0A182KXY3 T1P8B3 A0A1I8MFJ3 T1I7S8 A0A1I8P6R0 A0A1Y1LDQ6 A0A1Y1L923 A0A0R3NGA2 B5DWU2 A0A0M4F5M5 A0A1B6E6H9 B4G2I1 A0A3B0K3P3 D6WCY0 B3MT48 W8BB89 A0A1W4UIQ0 A0A1W4U5E1 A0A1W4UGV9 B4NBG4 B4LYI7 B4K8N1 A0A1J1I8E4 A0A195BIR1 Q7KRW4 A0A310SPN7 B4JTM7
A0A3S2PJF9 A0A2A4ITR7 E2BB79 K7INE5 E2ALT1 A0A1Q3FZ21 A0A1Q3FZ53 A0A084WR42 A0A182R8C8 A0A158P3F3 A0A182NCK3 A0A182GA80 A0A195F0B1 Q178P2 A0A182IV65 A0A026W0P7 A0A232EWK0 A0A3L8E4L7 A0A182YG74 A0A087ZUG8 A0A2A3EJS2 A0A182Q4G3 A0A182T1W9 A0A2J7RAG3 F5HLF5 A0A182F8E0 A0A0M9A6N6 A0A182PTT0 A0A0L7QWL2 A0A182KFB7 A0A182W2Z0 A0A182U9P3 A0A182VAQ1 A0A182WVF9 A0A182I938 W5JHD0 A0A195BIK5 A0A195DN33 A0A1S4FBS0 A0A1B0CGE3 A0A336MBJ6 A0A336M794 A0A336KRZ8 A0A336KP62 A0A1L8E2W2 A0A1L8E2E6 A0A067QSR5 A0A195CCY0 A0A151WZ01 A0A1B6CLC6 A0A0L0C6T3 A0A034V8K5 F4W4E5 A0A195DN37 E9JC17 A0A0K8UB09 E0VS71 A0A0A1WFD9 A0A0P4VSA7 A0A2R7VTS0 A0A069DXK6 A0A224X6F7 A0A154PMQ1 A0A1B6M3I9 A0A146LYB5 A0A0V0G738 A0A1B6GWM4 A0A182KXY3 T1P8B3 A0A1I8MFJ3 T1I7S8 A0A1I8P6R0 A0A1Y1LDQ6 A0A1Y1L923 A0A0R3NGA2 B5DWU2 A0A0M4F5M5 A0A1B6E6H9 B4G2I1 A0A3B0K3P3 D6WCY0 B3MT48 W8BB89 A0A1W4UIQ0 A0A1W4U5E1 A0A1W4UGV9 B4NBG4 B4LYI7 B4K8N1 A0A1J1I8E4 A0A195BIR1 Q7KRW4 A0A310SPN7 B4JTM7
PDB
5LDS
E-value=1.10025e-155,
Score=1415
Ontologies
PATHWAY
GO
PANTHER
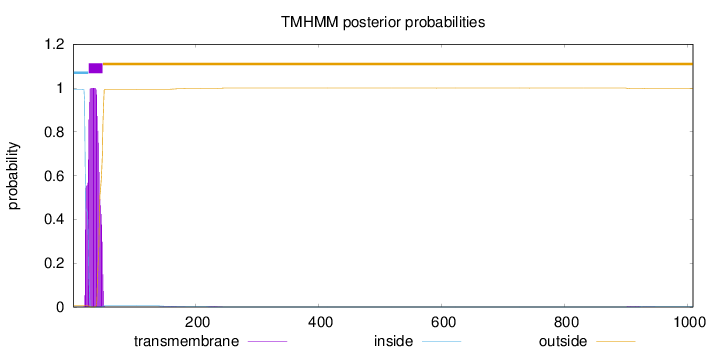
Topology
Length:
1009
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.53431
Exp number, first 60 AAs:
22.3693
Total prob of N-in:
0.99418
POSSIBLE N-term signal
sequence
inside
1 - 25
TMhelix
26 - 48
outside
49 - 1009
Population Genetic Test Statistics
Pi
169.673173
Theta
152.831283
Tajima's D
-1.519217
CLR
1.8843
CSRT
0.0580970951452427
Interpretation
Uncertain
