Gene
KWMTBOMO04884
Pre Gene Modal
BGIBMGA008019
Annotation
AF173553_1_V-ATPase_110_kDa_integral_membrane_subunit_[Manduca_sexta]
Full name
V-type proton ATPase subunit a
Location in the cell
PlasmaMembrane Reliability : 4.866
Sequence
CDS
ATGGGCTCCATGTTCAGGAGCGAGGAGATGGTCCTCTGTCAGCTCTTCGTTCAGCCGGAGGCGGCTTACGTCTCCATGTACGAGCTAGGAGAAGCTGGTATTGCTCAATTCCGAGATTTGAATCCACACGTAAATGACTTCCAAAGAAGATACGTAAGTGAAGTTCGCCGCTGCAGCGAGATGGAACGCAAGCTGCGATGGGTGAGTGGAGAGCTACCGGAGCCTCCTCCTCCGCCCATACCCTCGCGACGGATGCTCAGTCCACGAGAGATCAATATGCTAGAGGAAAAAATTGATTACATCGAATCAGAAATCCAAGAGATGACAAAAAACGCTCGAAACCTCAAAAGCGACTACCTTTCCTTGATAGAGCTGAAACTTCTCATAGAGAAAACACAGACCTTCTTCCAAGATCACAGCGCCCAGAGGAAACTGTCGGCCACCGTTCAGATCTACAACAATGACGCTGTTTTCGGGCACCTCGGCTTCATCGCGGGAACAGTGGCCACATCCAGGGTGCCTTCGTTTGAAAGGATGCTATGGAGGATATCGCACGGGAACATATTCTTCAAACAGGCACAGATCGATCAGCCTCTAAAAGATCCCATTACGGGTCACGAACTACAAAAGACTGTCTTCGTAGTATTCTTCCACGGTGAGCAGATCAAGTTGAGAGTGAAGAAGGTCTGCTTTGGGTTTAAAGCTACTCTGTACCCATGCCCTGCGACGTATAAGGACCAGCAAGAAATGCTGTCTGGAGTCGAAACGAGGATCAAGGACTTAGAAATGGTACTAGACCAGACAGAGCAGCACAGACGATTGGTCCTCACCAATATAGCAGGAGACATCAGCACGTGGATGCTGGCCGTAAGGAAGGAGAAGGCCATCTACCATACCCTCAACATGTTCAGCATGGACATCATTAAGAAATGCCTCATCGCTGAATGCTGGGTTCCAAAAAGTGATCTTTACATTGCCCAGAAGGCTCTGGATGATGGAGTGAAAGCAAGTGGCAGTCCCATTCCATCCATCCTCCACCACGTGCCAACCAGAGAAATACCACCCACTTTTAACAGAACCAACAAATTCACGAGGGGATTTCAAACTCTTATTGATTCCTATGGAATCGCCAGCTACAGAGAAGTAAATCCAGCGCTATACACAATCATAACATTCCCGTTTCTGTTCGCCGTCATGTTCGGCGACGTAGGCCATGGACTTATCATGACTGCCTTTTCTGCGATCATCGTTATATACGAGAAGAGACTCTCGAAGAATAAATCGGACAACGAAATATGGAACATTTTCTTTGCCGGACGATACATTATCTTACTGATGGGAATATTCTCTGTCTATACCGGTTTGATCTACAACGACATGTTCTCGAAGTCTCTGAATCTGTTCGGAAGTAGTTGGATGAACGTTTACGATAGTGAAACCCTTGCGAATCATACATCGTTGGAATTGGATCCGGCTGTGGCCTACACTCAAACGCCATATCCTTTCGGCTTGGACCCGGCGTGGCAGTTCGCCACGAACAACATAATATTTTTGAATTCATTCAAAATGAAACTGTCCATTATATTCGGCGTACTGCACATGGCGTTCGGTGTGACAATGAGTGTGGTAAACTTCAATTTCTTCAGGAAGCCGGAAATGATTTATCTCCAGTACGTGCCGCAAATTCTGTTTCTGATGCTGCTGTTCTGGTACCTCTGTATATTGATGTTCATGAAGTGGACCATGTACTCCGCCACGGCGACAGACCCGACATACGGTACGTCTTGTGCCCCATCAGTCCTAATAACCTTCATCAACATGATGCTGCTGAAGGGCTCTGAGGACAGCCCTCCGTGTAAGGCGATGATGTTCGACGGCCAGGACGCATTGCAGAAGTTATTCTTGACGCTTGGCTTCCTTTGCATTCCGATAATGTTATTCGGGAAACCGGCATTCTTGTTCTGGCAAGCCCGTAACCGGAAGCGCCACGAGGTGGGCGGGGAGCGCGGGGAGGTGCCGGCGGGGGCCGGCGGGGGTGCGGGGGCGGCGGGGGAGCTGCTGATCACGCAGGCGATACACACCATAGAGTACGTGCTGGGCACCGTCTCGCACACCGCCTCCTACCTGCGCCTCTGGGCCCTCTCCCTCGCGCACGCACAGCTGTCGGCCGTGCTGTGGCAGCGCGTGCTCCGCTACGGCCTGGCCAGGAGTCTGGCGGGCTCCGTCACCATGTACGTGGGGTTCGCGGTGTGGGCCTTCTTCACGCTCGCCATCCTCGTCCTCATGGAAGGACTGTCTGCCTTCCTGCACACGCTGCGGTTGCACTGGGTCGAGTTCATGAGTAAGTTCTACGACGGACAAGGATACGCTTTCATGCCGTTCTCATTCGCGGCCATCATAGAGCACGACGACGAGGAACTGGCGCAAAATGGCGCGCCGCCCGAGAAGAAAGACGAAGAATATTAA
Protein
MGSMFRSEEMVLCQLFVQPEAAYVSMYELGEAGIAQFRDLNPHVNDFQRRYVSEVRRCSEMERKLRWVSGELPEPPPPPIPSRRMLSPREINMLEEKIDYIESEIQEMTKNARNLKSDYLSLIELKLLIEKTQTFFQDHSAQRKLSATVQIYNNDAVFGHLGFIAGTVATSRVPSFERMLWRISHGNIFFKQAQIDQPLKDPITGHELQKTVFVVFFHGEQIKLRVKKVCFGFKATLYPCPATYKDQQEMLSGVETRIKDLEMVLDQTEQHRRLVLTNIAGDISTWMLAVRKEKAIYHTLNMFSMDIIKKCLIAECWVPKSDLYIAQKALDDGVKASGSPIPSILHHVPTREIPPTFNRTNKFTRGFQTLIDSYGIASYREVNPALYTIITFPFLFAVMFGDVGHGLIMTAFSAIIVIYEKRLSKNKSDNEIWNIFFAGRYIILLMGIFSVYTGLIYNDMFSKSLNLFGSSWMNVYDSETLANHTSLELDPAVAYTQTPYPFGLDPAWQFATNNIIFLNSFKMKLSIIFGVLHMAFGVTMSVVNFNFFRKPEMIYLQYVPQILFLMLLFWYLCILMFMKWTMYSATATDPTYGTSCAPSVLITFINMMLLKGSEDSPPCKAMMFDGQDALQKLFLTLGFLCIPIMLFGKPAFLFWQARNRKRHEVGGERGEVPAGAGGGAGAAGELLITQAIHTIEYVLGTVSHTASYLRLWALSLAHAQLSAVLWQRVLRYGLARSLAGSVTMYVGFAVWAFFTLAILVLMEGLSAFLHTLRLHWVEFMSKFYDGQGYAFMPFSFAAIIEHDDEELAQNGAPPEKKDEEY
Summary
Description
Essential component of the vacuolar proton pump (V-ATPase), a multimeric enzyme that catalyzes the translocation of protons across the membranes. Required for assembly and activity of the V-ATPase.
Similarity
Belongs to the V-ATPase 116 kDa subunit family.
Uniprot
Q9NJA4
A0A212EGQ2
A0A2W1BWJ3
A0A194QCV9
A0A194QKQ6
A0A2H1W9Q1
+ More
A0A194QD04 H9IYU7 A0A1E1WPT2 A0A194RMC5 A0A1B1UZP4 A0A212ESI2 A0A1Q3FCP1 B4QUE6 U5EY09 A0A2J7RKU2 A0A182R647 A0A2H8TPX0 A0A182QWU8 A0A182VZ95 A0A182PGR3 A0A182M9P0 B4I290 A0A2M4A171 A0A2M4A1G6 Q9VE75 A0A1W4U7R6 A0A182NS68 B4PLX2 A0A182IX69 B3LW99 A0A182WSF2 A0A2C9GQN9 A0A182L9N2 A0A182F7K3 Q7PKS7 A0A182USZ7 A0A182UDQ6 A0A3B0KN56 W5JMD3 A0A2M4BEH6 A0A023EW58 B4M3Z5 A0A1I8NRD1 Q299L9 A0A1L8DEM8 A0A2M3YYN2 B4G5D7 Q16HE2 A0A182YAM6 A0A1B6MKV2 A0A1L8DEX4 A0A2M3YYH4 A0A067RRK3 A0A0M5J4P9 A0A1B6MPW5 B4K928 Q8T5K2 T1PG42 A0A195CD36 B4N9D9 A0A0K8TQR7 D6WCX7 B4JF18 A0A1L8EDB7 A0A1L8EDJ5 A0A1J1HJB8 A0A0K8UV85 A0A195BIS9 A0A034VLS7 A0A087ZUG7 A0A336KNK4 A0A336M7G7 A0A1A9VER2 W8BF11 A0A1A9ZXN1 E0V9Q8 A0A1B0BJB3 A0A182K5C5 U4U2F9 N6TTR3 A0A182M1T3 V9ILL6 A0A0A9Z8V9 A0A182V6H6 A0A0A1X2U8 A0A0L7QWI4 A0A195DP30 A0A2C9GQN1 K7J2E3 A0A0A9ZE97 A0A0K8T8E8 A0A182YAM5 A0A1Y1JSJ5 A0A224X7A5 A0A182L9N3 A0A1Y1JVN1
A0A194QD04 H9IYU7 A0A1E1WPT2 A0A194RMC5 A0A1B1UZP4 A0A212ESI2 A0A1Q3FCP1 B4QUE6 U5EY09 A0A2J7RKU2 A0A182R647 A0A2H8TPX0 A0A182QWU8 A0A182VZ95 A0A182PGR3 A0A182M9P0 B4I290 A0A2M4A171 A0A2M4A1G6 Q9VE75 A0A1W4U7R6 A0A182NS68 B4PLX2 A0A182IX69 B3LW99 A0A182WSF2 A0A2C9GQN9 A0A182L9N2 A0A182F7K3 Q7PKS7 A0A182USZ7 A0A182UDQ6 A0A3B0KN56 W5JMD3 A0A2M4BEH6 A0A023EW58 B4M3Z5 A0A1I8NRD1 Q299L9 A0A1L8DEM8 A0A2M3YYN2 B4G5D7 Q16HE2 A0A182YAM6 A0A1B6MKV2 A0A1L8DEX4 A0A2M3YYH4 A0A067RRK3 A0A0M5J4P9 A0A1B6MPW5 B4K928 Q8T5K2 T1PG42 A0A195CD36 B4N9D9 A0A0K8TQR7 D6WCX7 B4JF18 A0A1L8EDB7 A0A1L8EDJ5 A0A1J1HJB8 A0A0K8UV85 A0A195BIS9 A0A034VLS7 A0A087ZUG7 A0A336KNK4 A0A336M7G7 A0A1A9VER2 W8BF11 A0A1A9ZXN1 E0V9Q8 A0A1B0BJB3 A0A182K5C5 U4U2F9 N6TTR3 A0A182M1T3 V9ILL6 A0A0A9Z8V9 A0A182V6H6 A0A0A1X2U8 A0A0L7QWI4 A0A195DP30 A0A2C9GQN1 K7J2E3 A0A0A9ZE97 A0A0K8T8E8 A0A182YAM5 A0A1Y1JSJ5 A0A224X7A5 A0A182L9N3 A0A1Y1JVN1
Pubmed
22118469
28756777
26354079
19121390
17994087
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 20966253 12364791 14747013 17210077 20920257 23761445 24945155 26483478 15632085 17510324 25244985 24845553 12060762 25315136 26369729 18362917 19820115 25348373 24495485 20566863 23537049 25401762 26823975 25830018 20075255 28004739
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 20966253 12364791 14747013 17210077 20920257 23761445 24945155 26483478 15632085 17510324 25244985 24845553 12060762 25315136 26369729 18362917 19820115 25348373 24495485 20566863 23537049 25401762 26823975 25830018 20075255 28004739
EMBL
AF173553
AAF28474.1
AGBW02015053
OWR40651.1
KZ149969
PZC76113.1
+ More
KQ459185 KPJ03378.1 KQ461198 KPJ05979.1 ODYU01006919 SOQ49194.1 KQ459220 KPJ02860.1 BABH01010757 BABH01010758 BABH01010759 GDQN01008557 GDQN01001991 JAT82497.1 JAT89063.1 KQ459984 KPJ18978.1 KU550964 ANW09710.1 AGBW02012778 OWR44452.1 GFDL01009700 JAV25345.1 CM000364 EDX12446.1 GANO01000631 JAB59240.1 NEVH01002707 PNF41457.1 GFXV01004235 MBW16040.1 AXCN02000267 AXCM01006747 CH480820 EDW54647.1 GGFK01001212 MBW34533.1 GGFK01001312 MBW34633.1 AF132183 AE014297 AAD34771.1 AAF55552.2 CM000160 EDW95904.1 CH902617 EDV42677.1 KPU79847.1 APCN01000888 AAAB01008987 EAA43151.2 OUUW01000013 SPP88009.1 ADMH02001150 ETN63909.1 GGFJ01002077 MBW51218.1 JXUM01123189 JXUM01123190 JXUM01123191 JXUM01123192 GAPW01000216 JAC13382.1 CH940652 EDW59356.1 KRF78813.1 CM000070 EAL27684.1 GFDF01009183 JAV04901.1 GGFM01000600 MBW21351.1 CH479179 EDW24803.1 CH478180 EAT33671.1 GEBQ01024009 GEBQ01003411 JAT15968.1 JAT36566.1 GFDF01009184 JAV04900.1 GGFM01000527 MBW21278.1 KK852470 KDR23260.1 CP012526 ALC46094.1 GEBQ01009678 GEBQ01002003 JAT30299.1 JAT37974.1 CH933806 EDW14441.1 KRG01017.1 AJ439060 CAD27758.1 KA646853 AFP61482.1 KQ977935 KYM98717.1 CH964232 EDW80572.1 GDAI01000904 JAI16699.1 KQ971311 EEZ99071.2 CH916369 EDV93299.1 GFDG01002069 JAV16730.1 GFDG01002067 JAV16732.1 CVRI01000006 CRK88011.1 GDHF01029529 GDHF01021876 JAI22785.1 JAI30438.1 KQ976467 KYM84195.1 GAKP01016222 JAC42730.1 UFQS01000635 UFQT01000635 SSX05612.1 SSX25971.1 SSX05611.1 SSX25970.1 GAMC01006625 JAB99930.1 DS234995 EEB10093.1 JXJN01015358 KB631625 ERL84786.1 APGK01052546 KB741213 ENN72620.1 AXCM01012792 JR053567 AEY61965.1 GBHO01001932 GDHC01015774 JAG41672.1 JAQ02855.1 GBXI01008638 JAD05654.1 KQ414710 KOC62988.1 KQ980713 KYN14244.1 AAZX01007551 GBHO01001936 GDHC01018959 JAG41668.1 JAP99669.1 GBRD01003998 JAG61823.1 GEZM01101879 JAV52239.1 GFTR01008140 JAW08286.1 GEZM01101880 GEZM01101878 JAV52238.1
KQ459185 KPJ03378.1 KQ461198 KPJ05979.1 ODYU01006919 SOQ49194.1 KQ459220 KPJ02860.1 BABH01010757 BABH01010758 BABH01010759 GDQN01008557 GDQN01001991 JAT82497.1 JAT89063.1 KQ459984 KPJ18978.1 KU550964 ANW09710.1 AGBW02012778 OWR44452.1 GFDL01009700 JAV25345.1 CM000364 EDX12446.1 GANO01000631 JAB59240.1 NEVH01002707 PNF41457.1 GFXV01004235 MBW16040.1 AXCN02000267 AXCM01006747 CH480820 EDW54647.1 GGFK01001212 MBW34533.1 GGFK01001312 MBW34633.1 AF132183 AE014297 AAD34771.1 AAF55552.2 CM000160 EDW95904.1 CH902617 EDV42677.1 KPU79847.1 APCN01000888 AAAB01008987 EAA43151.2 OUUW01000013 SPP88009.1 ADMH02001150 ETN63909.1 GGFJ01002077 MBW51218.1 JXUM01123189 JXUM01123190 JXUM01123191 JXUM01123192 GAPW01000216 JAC13382.1 CH940652 EDW59356.1 KRF78813.1 CM000070 EAL27684.1 GFDF01009183 JAV04901.1 GGFM01000600 MBW21351.1 CH479179 EDW24803.1 CH478180 EAT33671.1 GEBQ01024009 GEBQ01003411 JAT15968.1 JAT36566.1 GFDF01009184 JAV04900.1 GGFM01000527 MBW21278.1 KK852470 KDR23260.1 CP012526 ALC46094.1 GEBQ01009678 GEBQ01002003 JAT30299.1 JAT37974.1 CH933806 EDW14441.1 KRG01017.1 AJ439060 CAD27758.1 KA646853 AFP61482.1 KQ977935 KYM98717.1 CH964232 EDW80572.1 GDAI01000904 JAI16699.1 KQ971311 EEZ99071.2 CH916369 EDV93299.1 GFDG01002069 JAV16730.1 GFDG01002067 JAV16732.1 CVRI01000006 CRK88011.1 GDHF01029529 GDHF01021876 JAI22785.1 JAI30438.1 KQ976467 KYM84195.1 GAKP01016222 JAC42730.1 UFQS01000635 UFQT01000635 SSX05612.1 SSX25971.1 SSX05611.1 SSX25970.1 GAMC01006625 JAB99930.1 DS234995 EEB10093.1 JXJN01015358 KB631625 ERL84786.1 APGK01052546 KB741213 ENN72620.1 AXCM01012792 JR053567 AEY61965.1 GBHO01001932 GDHC01015774 JAG41672.1 JAQ02855.1 GBXI01008638 JAD05654.1 KQ414710 KOC62988.1 KQ980713 KYN14244.1 AAZX01007551 GBHO01001936 GDHC01018959 JAG41668.1 JAP99669.1 GBRD01003998 JAG61823.1 GEZM01101879 JAV52239.1 GFTR01008140 JAW08286.1 GEZM01101880 GEZM01101878 JAV52238.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000005204
UP000000304
UP000235965
+ More
UP000075886 UP000075920 UP000075885 UP000075883 UP000001292 UP000000803 UP000192221 UP000075884 UP000002282 UP000075880 UP000007801 UP000076407 UP000075840 UP000075882 UP000069272 UP000007062 UP000075903 UP000075902 UP000268350 UP000000673 UP000069940 UP000008792 UP000095300 UP000001819 UP000008744 UP000008820 UP000076408 UP000027135 UP000092553 UP000009192 UP000095301 UP000078542 UP000007798 UP000007266 UP000001070 UP000183832 UP000078540 UP000005203 UP000078200 UP000092445 UP000009046 UP000092460 UP000075881 UP000030742 UP000019118 UP000053825 UP000078492 UP000002358
UP000075886 UP000075920 UP000075885 UP000075883 UP000001292 UP000000803 UP000192221 UP000075884 UP000002282 UP000075880 UP000007801 UP000076407 UP000075840 UP000075882 UP000069272 UP000007062 UP000075903 UP000075902 UP000268350 UP000000673 UP000069940 UP000008792 UP000095300 UP000001819 UP000008744 UP000008820 UP000076408 UP000027135 UP000092553 UP000009192 UP000095301 UP000078542 UP000007798 UP000007266 UP000001070 UP000183832 UP000078540 UP000005203 UP000078200 UP000092445 UP000009046 UP000092460 UP000075881 UP000030742 UP000019118 UP000053825 UP000078492 UP000002358
Pfam
PF01496 V_ATPase_I
ProteinModelPortal
Q9NJA4
A0A212EGQ2
A0A2W1BWJ3
A0A194QCV9
A0A194QKQ6
A0A2H1W9Q1
+ More
A0A194QD04 H9IYU7 A0A1E1WPT2 A0A194RMC5 A0A1B1UZP4 A0A212ESI2 A0A1Q3FCP1 B4QUE6 U5EY09 A0A2J7RKU2 A0A182R647 A0A2H8TPX0 A0A182QWU8 A0A182VZ95 A0A182PGR3 A0A182M9P0 B4I290 A0A2M4A171 A0A2M4A1G6 Q9VE75 A0A1W4U7R6 A0A182NS68 B4PLX2 A0A182IX69 B3LW99 A0A182WSF2 A0A2C9GQN9 A0A182L9N2 A0A182F7K3 Q7PKS7 A0A182USZ7 A0A182UDQ6 A0A3B0KN56 W5JMD3 A0A2M4BEH6 A0A023EW58 B4M3Z5 A0A1I8NRD1 Q299L9 A0A1L8DEM8 A0A2M3YYN2 B4G5D7 Q16HE2 A0A182YAM6 A0A1B6MKV2 A0A1L8DEX4 A0A2M3YYH4 A0A067RRK3 A0A0M5J4P9 A0A1B6MPW5 B4K928 Q8T5K2 T1PG42 A0A195CD36 B4N9D9 A0A0K8TQR7 D6WCX7 B4JF18 A0A1L8EDB7 A0A1L8EDJ5 A0A1J1HJB8 A0A0K8UV85 A0A195BIS9 A0A034VLS7 A0A087ZUG7 A0A336KNK4 A0A336M7G7 A0A1A9VER2 W8BF11 A0A1A9ZXN1 E0V9Q8 A0A1B0BJB3 A0A182K5C5 U4U2F9 N6TTR3 A0A182M1T3 V9ILL6 A0A0A9Z8V9 A0A182V6H6 A0A0A1X2U8 A0A0L7QWI4 A0A195DP30 A0A2C9GQN1 K7J2E3 A0A0A9ZE97 A0A0K8T8E8 A0A182YAM5 A0A1Y1JSJ5 A0A224X7A5 A0A182L9N3 A0A1Y1JVN1
A0A194QD04 H9IYU7 A0A1E1WPT2 A0A194RMC5 A0A1B1UZP4 A0A212ESI2 A0A1Q3FCP1 B4QUE6 U5EY09 A0A2J7RKU2 A0A182R647 A0A2H8TPX0 A0A182QWU8 A0A182VZ95 A0A182PGR3 A0A182M9P0 B4I290 A0A2M4A171 A0A2M4A1G6 Q9VE75 A0A1W4U7R6 A0A182NS68 B4PLX2 A0A182IX69 B3LW99 A0A182WSF2 A0A2C9GQN9 A0A182L9N2 A0A182F7K3 Q7PKS7 A0A182USZ7 A0A182UDQ6 A0A3B0KN56 W5JMD3 A0A2M4BEH6 A0A023EW58 B4M3Z5 A0A1I8NRD1 Q299L9 A0A1L8DEM8 A0A2M3YYN2 B4G5D7 Q16HE2 A0A182YAM6 A0A1B6MKV2 A0A1L8DEX4 A0A2M3YYH4 A0A067RRK3 A0A0M5J4P9 A0A1B6MPW5 B4K928 Q8T5K2 T1PG42 A0A195CD36 B4N9D9 A0A0K8TQR7 D6WCX7 B4JF18 A0A1L8EDB7 A0A1L8EDJ5 A0A1J1HJB8 A0A0K8UV85 A0A195BIS9 A0A034VLS7 A0A087ZUG7 A0A336KNK4 A0A336M7G7 A0A1A9VER2 W8BF11 A0A1A9ZXN1 E0V9Q8 A0A1B0BJB3 A0A182K5C5 U4U2F9 N6TTR3 A0A182M1T3 V9ILL6 A0A0A9Z8V9 A0A182V6H6 A0A0A1X2U8 A0A0L7QWI4 A0A195DP30 A0A2C9GQN1 K7J2E3 A0A0A9ZE97 A0A0K8T8E8 A0A182YAM5 A0A1Y1JSJ5 A0A224X7A5 A0A182L9N3 A0A1Y1JVN1
PDB
6C6L
E-value=5.35497e-128,
Score=1175
Ontologies
PATHWAY
GO
PANTHER
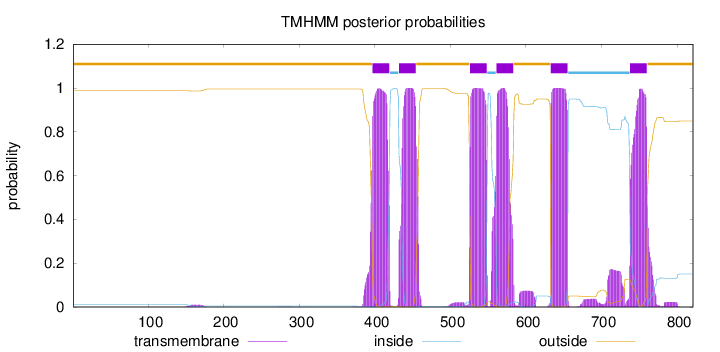
Topology
Length:
821
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
144.13707
Exp number, first 60 AAs:
0.00137
Total prob of N-in:
0.01290
outside
1 - 396
TMhelix
397 - 419
inside
420 - 431
TMhelix
432 - 454
outside
455 - 525
TMhelix
526 - 548
inside
549 - 560
TMhelix
561 - 583
outside
584 - 632
TMhelix
633 - 655
inside
656 - 737
TMhelix
738 - 760
outside
761 - 821
Population Genetic Test Statistics
Pi
198.476418
Theta
161.034872
Tajima's D
0.767026
CLR
0.137366
CSRT
0.599570021498925
Interpretation
Uncertain
