Gene
KWMTBOMO04883
Pre Gene Modal
BGIBMGA008056
Annotation
PREDICTED:_katanin_p60_ATPase-containing_subunit_A-like_1_[Bombyx_mori]
Full name
Katanin p60 ATPase-containing subunit A1
Alternative Name
p60 katanin
Location in the cell
Mitochondrial Reliability : 1.795 Nuclear Reliability : 2.068
Sequence
CDS
ATGACGAATGACCTGAAAGCTTCAACGAGCGAGAAGGAGGCACGGGGCTCCTGGCCGAGGAGGGACGACCTCAAGATTGAGTTCGGGTTCGGAATGCCTCTAGGGGCCATGAGTGCTCATCCAGCTTTCCACTCCGCTTGGGACATCGCTTGCCCATCTTCAGGCTTTCATCAGATGGCTCGTATGATGGACCGTATGATGGTGGAGAGCCTCCAGCCGTTTGGCTTCGCTAGGATGACGGCAAGACAGCGTGGTGATAGTCGTGATCATCAGCCTCCACGTGATGGTCTACAGACAGAGCGCCCTGATACGAGGAAGGAGCGGGCGAAGAATCATTCCAATACGCACGCCGCCACTGGTACCAGACATCCTCCTACACAAATAGTGAAACCAACTCAGTCGAATACATTTCTAGATCAGTCACAAGAGCAAAATAAACAAACGGACCCCACGACTGAGAAATGGGCGGGATCGTTACGGAAACGCGACCCTGACATCCAGCCGACCTTGCCGTCCATTGTCGCTAGAAAGACCTCTTCATCTTCAGTGGGCAGTGGAAGGAAGACCAGGTCTGTGGAAAGGATGCCCCGACAACGTGTAAGGCAGTTGGCTCCAATTAAACCGATTGTTAGAAAGGCTGAAGCGTCAGAAGGACAAGATGTTACAATTTATGAGAAGGATGAAAAAACATTCGATGCGACCGGCTATGAAATGCATCTAGTGGAGACCCTAGAGAGAGATATATTGCAGAGGAATCCTGATGTGCGCTGGAAGGATGTCATTGGATTGGACGATGCAAAAGCCGTGCTACAAGAGGCGATGGTCCTGCCTTTAGTGATGCCTGATTACTTTAAGGGTATAAGGAGACCTTGGAAGGGCGTGCTCCTCACAGGACCTCCGGGTACAGGGAAGACATTGCTCGCGAGGGCTGTGGCCACAGAGTGTAGAACCACTTTCTTTAATGTATCTTCGGCCACACTCACTTCGAAGTACAGGGGAGATTCTGAGAAATTGGTCAGACTTCTTTTTGACATGGCGTCTTTCTACGCGCCCAGCACGATCTTCTTGGACGAGGTGGACTCATTGTGCGCGGTCCGCGGTGCTGACTCCGAGCACGAGGCCTCGCGACGATTCAAAGCAGAGCTGCTCATACAAATGGACGGGCTTGCTGCCGTATTCAACCAAGATAAAATCATAATGGTATTAGCCGCGACCAACCATCCGTGGGACATCGACGAAGCCTTCAGAAGGAGGTTTGAGAAGAGGATCTACATTGGCCTTCCAGATGAGCCGACGCGTGTGAAACTTCTGAAGCTGTGTCTCCGAGAAGTCGCGCTAGCGGAAGATGTAGATCTAGTGGATCTGGCAACTAAATTGGACGGTTATAGCGGATCTGACATCTGCAACTTGTGCCGGGATGCCGCAATGATGACGATGCGTCGTAAGATAGCCGGTAAGAGTCCGGATCAGATCCGCAGACTGAAGCGCTCCGAGCTGGAGGCGCCGGTGTCGGCGGCCGACCTGGGCGCCTCCGCGGACAAGACGCGGCGCACGGTGAGCCCCGCCGACGTGGCGCGCTACTCCTCCTGGATGCAGCGCCACGGCTGCTCCTAG
Protein
MTNDLKASTSEKEARGSWPRRDDLKIEFGFGMPLGAMSAHPAFHSAWDIACPSSGFHQMARMMDRMMVESLQPFGFARMTARQRGDSRDHQPPRDGLQTERPDTRKERAKNHSNTHAATGTRHPPTQIVKPTQSNTFLDQSQEQNKQTDPTTEKWAGSLRKRDPDIQPTLPSIVARKTSSSSVGSGRKTRSVERMPRQRVRQLAPIKPIVRKAEASEGQDVTIYEKDEKTFDATGYEMHLVETLERDILQRNPDVRWKDVIGLDDAKAVLQEAMVLPLVMPDYFKGIRRPWKGVLLTGPPGTGKTLLARAVATECRTTFFNVSSATLTSKYRGDSEKLVRLLFDMASFYAPSTIFLDEVDSLCAVRGADSEHEASRRFKAELLIQMDGLAAVFNQDKIIMVLAATNHPWDIDEAFRRRFEKRIYIGLPDEPTRVKLLKLCLREVALAEDVDLVDLATKLDGYSGSDICNLCRDAAMMTMRRKIAGKSPDQIRRLKRSELEAPVSAADLGASADKTRRTVSPADVARYSSWMQRHGCS
Summary
Description
Catalytic subunit of a complex which severs microtubules in an ATP-dependent manner. Microtubule severing may promote rapid reorganization of cellular microtubule arrays and the release of microtubules from the centrosome following nucleation.
Catalytic Activity
n ATP + n H(2)O + a microtubule = n ADP + n phosphate + (n+1) alpha/beta tubulin heterodimers.
Subunit
Can homooligomerize into hexameric rings, which may be promoted by interaction with microtubules. Interacts with KATNB1, which may serve as a targeting subunit.
Similarity
Belongs to the AAA ATPase family. Katanin p60 subunit A1 subfamily.
Belongs to the CRISP family.
Belongs to the AAA ATPase family.
Belongs to the CRISP family.
Belongs to the AAA ATPase family.
Uniprot
A0A3S2NKC8
A0A194QD82
A0A194QR23
H9JEV9
A0A212EGP7
A0A2W1BM10
+ More
A0A2H1W7X2 A0A0L7LVK5 A0A1B6CIC8 B0W341 Q177C8 A0A1B6CXW1 A0A0L0CD63 T1HME4 A0A1B0GEC0 A0A1I8NSQ3 A0A336L4J8 A0A1B0GH48 B3LWD9 A0A2R7X9S4 A0A0P8YGG1 A0A0Q9WG34 A0A1B6HMU4 A0A336MBH7 A0A1B0DLX7 A0A336M4T2 A0A1B6HDV7 Q9VNI0 A0A1I8MC48 A0A0Q9WUU4 A0A0R1E714 B4QXK0 B4NJI3 A0A023F3Q6 A0A0K8VXN7 A0A034VCA9 A0A1B0C2K0 A0A1I8MC52 A0A1W4UYG0 A0A1S4FCZ1 Q4QPP5 A0A0R1E871 A0A1A9X217 B4KAH2 B4I474 A0A1A9YJT2 A0A034VCB3 B4PUY2 B3P2K0 B4G498 A0A224XFB1 B5DWF9 A0A1B0AA24 A0A1W4UJS3 A0A1A9VH43 A0A0M4EPI3 A0A0Q9X0L6 A0A1I8NSQ9 A0A0R3NHU4 A0A0P9A166 A0A0P8Y105 B4LYP5 A0A0A1WMX6 A0A182TIC9 A0A182WYZ4 A0A1B6FQN3 A0A182HJC0 A0A182VQQ6 A0A0Q9WS09 A0A182VL05 B4JG93 A0A182K733 A0A182N587 A0A1B6GCI6 A0A182MKQ4 Q961H1 A0A182RTT0 W8B3K0 A0A3F2Z047 A0A182ITX1 A0A1W4UWS4 A0A0Q9WUU5 E1JJ62 A0A1S3D1F0 A0A0R1E8U3 E0W309 A0A182GX67 A0A2C9NGQ7 A0A0Q9X4R0 A0A182FSJ0 A0A0Q9XB01 A0A1W4UX28 A0A1W4UJA6 A0A0R1E405 A0A182PE66 A0A182Q4L5 A0A0Q9X0P7 A0A1J1IJA5 I5AP40 A0A0Q9XAR9
A0A2H1W7X2 A0A0L7LVK5 A0A1B6CIC8 B0W341 Q177C8 A0A1B6CXW1 A0A0L0CD63 T1HME4 A0A1B0GEC0 A0A1I8NSQ3 A0A336L4J8 A0A1B0GH48 B3LWD9 A0A2R7X9S4 A0A0P8YGG1 A0A0Q9WG34 A0A1B6HMU4 A0A336MBH7 A0A1B0DLX7 A0A336M4T2 A0A1B6HDV7 Q9VNI0 A0A1I8MC48 A0A0Q9WUU4 A0A0R1E714 B4QXK0 B4NJI3 A0A023F3Q6 A0A0K8VXN7 A0A034VCA9 A0A1B0C2K0 A0A1I8MC52 A0A1W4UYG0 A0A1S4FCZ1 Q4QPP5 A0A0R1E871 A0A1A9X217 B4KAH2 B4I474 A0A1A9YJT2 A0A034VCB3 B4PUY2 B3P2K0 B4G498 A0A224XFB1 B5DWF9 A0A1B0AA24 A0A1W4UJS3 A0A1A9VH43 A0A0M4EPI3 A0A0Q9X0L6 A0A1I8NSQ9 A0A0R3NHU4 A0A0P9A166 A0A0P8Y105 B4LYP5 A0A0A1WMX6 A0A182TIC9 A0A182WYZ4 A0A1B6FQN3 A0A182HJC0 A0A182VQQ6 A0A0Q9WS09 A0A182VL05 B4JG93 A0A182K733 A0A182N587 A0A1B6GCI6 A0A182MKQ4 Q961H1 A0A182RTT0 W8B3K0 A0A3F2Z047 A0A182ITX1 A0A1W4UWS4 A0A0Q9WUU5 E1JJ62 A0A1S3D1F0 A0A0R1E8U3 E0W309 A0A182GX67 A0A2C9NGQ7 A0A0Q9X4R0 A0A182FSJ0 A0A0Q9XB01 A0A1W4UX28 A0A1W4UJA6 A0A0R1E405 A0A182PE66 A0A182Q4L5 A0A0Q9X0P7 A0A1J1IJA5 I5AP40 A0A0Q9XAR9
EC Number
5.6.1.1
Pubmed
EMBL
RSAL01000014
RVE53198.1
KQ459185
KPJ03379.1
KQ461198
KPJ05981.1
+ More
BABH01023380 BABH01023381 AGBW02015053 OWR40650.1 KZ149969 PZC76112.1 ODYU01006919 SOQ49195.1 JTDY01000015 KOB79414.1 GEDC01024072 JAS13226.1 DS231830 EDS30749.1 CH477378 EAT42264.1 GEDC01019133 JAS18165.1 JRES01000562 KNC30181.1 ACPB03001254 CCAG010012182 UFQS01001826 UFQT01001826 SSX12470.1 SSX31913.1 AJWK01003967 AJWK01003968 AJWK01003969 CH902617 EDV43772.1 KK854002 PTY32650.1 KPU80448.1 CH940650 KRF83030.1 GECU01031693 JAS76013.1 UFQT01000344 SSX23408.1 AJVK01016591 AJVK01016592 SSX23407.1 GECU01034836 JAS72870.1 AE014297 BT001274 AAF51954.1 AAN71030.1 CH964272 KRF99918.1 CM000160 KRK03971.1 CM000364 EDX11786.1 EDW83907.1 GBBI01003183 JAC15529.1 GDHF01009019 JAI43295.1 GAKP01018006 GAKP01018004 JAC40946.1 JXJN01024565 BT023721 AAY85121.1 ACZ94822.1 KRK03969.1 CH933806 EDW15685.2 CH480821 EDW55017.1 GAKP01018001 JAC40951.1 EDW97789.1 CH954181 EDV47950.1 CH479179 EDW25136.1 GFTR01006712 JAW09714.1 CM000070 EDY68150.1 CP012526 ALC46764.1 KRG01679.1 KRT00558.1 KPU80447.1 KPU80449.1 EDW66972.1 GBXI01014524 JAC99767.1 GECZ01017247 JAS52522.1 APCN01002144 APCN01002145 APCN01002146 KRF83029.1 CH916369 EDV93660.1 GECZ01016882 GECZ01009623 JAS52887.1 JAS60146.1 AXCM01005290 AY051591 AAF51955.2 AAK93015.1 GAMC01013413 GAMC01013412 JAB93142.1 KRF99919.1 ACZ94821.1 KRK03970.1 DS235881 EEB20015.1 JXUM01094777 JXUM01094778 JXUM01094779 JXUM01094780 JXUM01094781 JXUM01094782 KQ564152 KXJ72677.1 AAZO01007315 KRF99917.1 KRG01681.1 KRK03968.1 AXCN02000164 KRG01680.1 CVRI01000047 CRK98521.1 EIM52725.1 KRG01678.1
BABH01023380 BABH01023381 AGBW02015053 OWR40650.1 KZ149969 PZC76112.1 ODYU01006919 SOQ49195.1 JTDY01000015 KOB79414.1 GEDC01024072 JAS13226.1 DS231830 EDS30749.1 CH477378 EAT42264.1 GEDC01019133 JAS18165.1 JRES01000562 KNC30181.1 ACPB03001254 CCAG010012182 UFQS01001826 UFQT01001826 SSX12470.1 SSX31913.1 AJWK01003967 AJWK01003968 AJWK01003969 CH902617 EDV43772.1 KK854002 PTY32650.1 KPU80448.1 CH940650 KRF83030.1 GECU01031693 JAS76013.1 UFQT01000344 SSX23408.1 AJVK01016591 AJVK01016592 SSX23407.1 GECU01034836 JAS72870.1 AE014297 BT001274 AAF51954.1 AAN71030.1 CH964272 KRF99918.1 CM000160 KRK03971.1 CM000364 EDX11786.1 EDW83907.1 GBBI01003183 JAC15529.1 GDHF01009019 JAI43295.1 GAKP01018006 GAKP01018004 JAC40946.1 JXJN01024565 BT023721 AAY85121.1 ACZ94822.1 KRK03969.1 CH933806 EDW15685.2 CH480821 EDW55017.1 GAKP01018001 JAC40951.1 EDW97789.1 CH954181 EDV47950.1 CH479179 EDW25136.1 GFTR01006712 JAW09714.1 CM000070 EDY68150.1 CP012526 ALC46764.1 KRG01679.1 KRT00558.1 KPU80447.1 KPU80449.1 EDW66972.1 GBXI01014524 JAC99767.1 GECZ01017247 JAS52522.1 APCN01002144 APCN01002145 APCN01002146 KRF83029.1 CH916369 EDV93660.1 GECZ01016882 GECZ01009623 JAS52887.1 JAS60146.1 AXCM01005290 AY051591 AAF51955.2 AAK93015.1 GAMC01013413 GAMC01013412 JAB93142.1 KRF99919.1 ACZ94821.1 KRK03970.1 DS235881 EEB20015.1 JXUM01094777 JXUM01094778 JXUM01094779 JXUM01094780 JXUM01094781 JXUM01094782 KQ564152 KXJ72677.1 AAZO01007315 KRF99917.1 KRG01681.1 KRK03968.1 AXCN02000164 KRG01680.1 CVRI01000047 CRK98521.1 EIM52725.1 KRG01678.1
Proteomes
UP000283053
UP000053268
UP000053240
UP000005204
UP000007151
UP000037510
+ More
UP000002320 UP000008820 UP000037069 UP000015103 UP000092444 UP000095300 UP000092461 UP000007801 UP000008792 UP000092462 UP000000803 UP000095301 UP000007798 UP000002282 UP000000304 UP000092460 UP000192221 UP000091820 UP000009192 UP000001292 UP000092443 UP000008711 UP000008744 UP000001819 UP000092445 UP000078200 UP000092553 UP000075902 UP000076407 UP000075840 UP000075920 UP000075903 UP000001070 UP000075881 UP000075884 UP000075883 UP000075900 UP000075880 UP000079169 UP000009046 UP000069940 UP000249989 UP000069272 UP000075885 UP000075886 UP000183832
UP000002320 UP000008820 UP000037069 UP000015103 UP000092444 UP000095300 UP000092461 UP000007801 UP000008792 UP000092462 UP000000803 UP000095301 UP000007798 UP000002282 UP000000304 UP000092460 UP000192221 UP000091820 UP000009192 UP000001292 UP000092443 UP000008711 UP000008744 UP000001819 UP000092445 UP000078200 UP000092553 UP000075902 UP000076407 UP000075840 UP000075920 UP000075903 UP000001070 UP000075881 UP000075884 UP000075883 UP000075900 UP000075880 UP000079169 UP000009046 UP000069940 UP000249989 UP000069272 UP000075885 UP000075886 UP000183832
Interpro
Gene 3D
ProteinModelPortal
A0A3S2NKC8
A0A194QD82
A0A194QR23
H9JEV9
A0A212EGP7
A0A2W1BM10
+ More
A0A2H1W7X2 A0A0L7LVK5 A0A1B6CIC8 B0W341 Q177C8 A0A1B6CXW1 A0A0L0CD63 T1HME4 A0A1B0GEC0 A0A1I8NSQ3 A0A336L4J8 A0A1B0GH48 B3LWD9 A0A2R7X9S4 A0A0P8YGG1 A0A0Q9WG34 A0A1B6HMU4 A0A336MBH7 A0A1B0DLX7 A0A336M4T2 A0A1B6HDV7 Q9VNI0 A0A1I8MC48 A0A0Q9WUU4 A0A0R1E714 B4QXK0 B4NJI3 A0A023F3Q6 A0A0K8VXN7 A0A034VCA9 A0A1B0C2K0 A0A1I8MC52 A0A1W4UYG0 A0A1S4FCZ1 Q4QPP5 A0A0R1E871 A0A1A9X217 B4KAH2 B4I474 A0A1A9YJT2 A0A034VCB3 B4PUY2 B3P2K0 B4G498 A0A224XFB1 B5DWF9 A0A1B0AA24 A0A1W4UJS3 A0A1A9VH43 A0A0M4EPI3 A0A0Q9X0L6 A0A1I8NSQ9 A0A0R3NHU4 A0A0P9A166 A0A0P8Y105 B4LYP5 A0A0A1WMX6 A0A182TIC9 A0A182WYZ4 A0A1B6FQN3 A0A182HJC0 A0A182VQQ6 A0A0Q9WS09 A0A182VL05 B4JG93 A0A182K733 A0A182N587 A0A1B6GCI6 A0A182MKQ4 Q961H1 A0A182RTT0 W8B3K0 A0A3F2Z047 A0A182ITX1 A0A1W4UWS4 A0A0Q9WUU5 E1JJ62 A0A1S3D1F0 A0A0R1E8U3 E0W309 A0A182GX67 A0A2C9NGQ7 A0A0Q9X4R0 A0A182FSJ0 A0A0Q9XB01 A0A1W4UX28 A0A1W4UJA6 A0A0R1E405 A0A182PE66 A0A182Q4L5 A0A0Q9X0P7 A0A1J1IJA5 I5AP40 A0A0Q9XAR9
A0A2H1W7X2 A0A0L7LVK5 A0A1B6CIC8 B0W341 Q177C8 A0A1B6CXW1 A0A0L0CD63 T1HME4 A0A1B0GEC0 A0A1I8NSQ3 A0A336L4J8 A0A1B0GH48 B3LWD9 A0A2R7X9S4 A0A0P8YGG1 A0A0Q9WG34 A0A1B6HMU4 A0A336MBH7 A0A1B0DLX7 A0A336M4T2 A0A1B6HDV7 Q9VNI0 A0A1I8MC48 A0A0Q9WUU4 A0A0R1E714 B4QXK0 B4NJI3 A0A023F3Q6 A0A0K8VXN7 A0A034VCA9 A0A1B0C2K0 A0A1I8MC52 A0A1W4UYG0 A0A1S4FCZ1 Q4QPP5 A0A0R1E871 A0A1A9X217 B4KAH2 B4I474 A0A1A9YJT2 A0A034VCB3 B4PUY2 B3P2K0 B4G498 A0A224XFB1 B5DWF9 A0A1B0AA24 A0A1W4UJS3 A0A1A9VH43 A0A0M4EPI3 A0A0Q9X0L6 A0A1I8NSQ9 A0A0R3NHU4 A0A0P9A166 A0A0P8Y105 B4LYP5 A0A0A1WMX6 A0A182TIC9 A0A182WYZ4 A0A1B6FQN3 A0A182HJC0 A0A182VQQ6 A0A0Q9WS09 A0A182VL05 B4JG93 A0A182K733 A0A182N587 A0A1B6GCI6 A0A182MKQ4 Q961H1 A0A182RTT0 W8B3K0 A0A3F2Z047 A0A182ITX1 A0A1W4UWS4 A0A0Q9WUU5 E1JJ62 A0A1S3D1F0 A0A0R1E8U3 E0W309 A0A182GX67 A0A2C9NGQ7 A0A0Q9X4R0 A0A182FSJ0 A0A0Q9XB01 A0A1W4UX28 A0A1W4UJA6 A0A0R1E405 A0A182PE66 A0A182Q4L5 A0A0Q9X0P7 A0A1J1IJA5 I5AP40 A0A0Q9XAR9
PDB
5ZQM
E-value=3.63052e-107,
Score=994
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
Cytoskeleton Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
Microtubule organizing center Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
Centrosome Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
Spindle pole Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
Spindle Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
Cytoskeleton Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
Microtubule organizing center Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
Centrosome Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
Spindle pole Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
Spindle Predominantly cytoplasmic. Also localized to the interphase centrosome and the mitotic spindle poles. Enhanced recruitment to the mitotic spindle poles requires microtubules and interaction with KATNB1. With evidence from 1 publications.
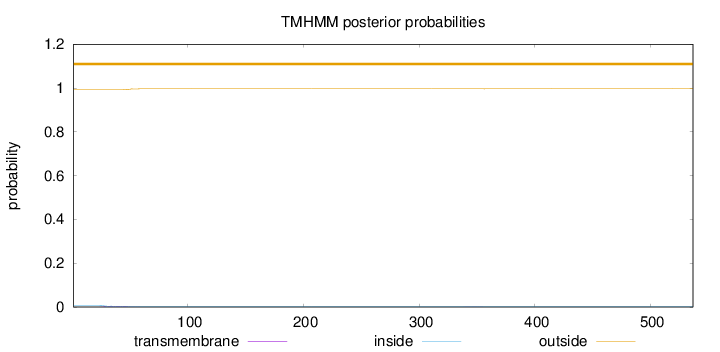
Length:
537
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10163
Exp number, first 60 AAs:
0.09688
Total prob of N-in:
0.00791
outside
1 - 537
Population Genetic Test Statistics
Pi
201.965495
Theta
162.797953
Tajima's D
0.724359
CLR
0.000001
CSRT
0.577321133943303
Interpretation
Uncertain
