Pre Gene Modal
BGIBMGA008053
Annotation
PREDICTED:_sphingosine-1-phosphate_lyase_[Bombyx_mori]
Full name
Sphingosine-1-phosphate lyase
Alternative Name
Sphingosine-1-phosphate aldolase
Location in the cell
Mitochondrial Reliability : 1.346 Peroxisomal Reliability : 1.523
Sequence
CDS
ATGAGTGAGAGACCACAGCCCTTAAGGGCTATAAACAAGTTTTTTGATGGAAAGGAGCCATGGCAAATTGTATCAATGACAGCTTCTTCTTTATTAGCCATAGTTTGGGTTCACAGTTTATACAATGCAAGAGAAAGTGTGACAGTCCGCCTTCGAAAACAGTTTTTCAGGTGGCTACGTCAAGTTCCAATGGTAAGGAAGAAGATAGAACAAAAAATGGCCGAGATAAAGAAAGATTTCCATACAGATGTAGAAAAGAGACTCGCAGGAGTCACAATTAGAAGAGAATTGCCTGAACAAGGTTTGGATCCTGAACAGATATCGCAAGAAGTCAGAGATCATCTGAATTTAGGGGCTTATGATTGGAAGAATGGTGCAGTATCTGGTGCAGTCTACCACTTGTGTGAAGACATCAGTCGTGTGGCTTGCGAGGCTTATACCTTGACTGCCTACACCAATCCACTGCACTCAGATGTCTTCCCAGGAATTAACAAAATGGAGGCGGAGATTGTGCAGATGGCTATTAACTTATTCAATGGTGATGAGGAGTGTTGTGGGACAGTAACTACAGGTGGCACAGAATCCATAATAATGGCGTGCAAGGCCTTCAGGGACCTCGCCTACAGCAAAGGCATAAGCAACCCACAGATAATCGTGCCATCAACTGCCCATTCTGCTTTCGACAAGGCGGCTCAGTACTTAGGCCTGTTTGTTGTGACCATACCGGTGGACCCGGTGACGCTATGTGTCGATGTCGATAAGGTCAGCGATGCTATTGGAAGACGAACGTGCTTGATCGTGGGGTCGGCTCCGAACTACCCGTACGGCACGATGGACGACATCGGCGCGTTGTCTGCGCTGGCGGCGCGCGCGGGGGTGCCGCTGCACGTGGACGCGTGCCTGGGCGGGCTGGTGGCGGGCTTCATGCCGGCCGCCGGACACCCCGTGCCGCCCTTCGACTTCCGCCTGCCCGGAGTCGCCAGCATCTCCGCCGACACGCACAAGTACGGGTACGCCCCGAAGGGCACGTCGGTGATCCTGTACCGGCGTGCGGAGTACCGGCACTGCCAGTACACCGTCACCACGGAGTGGCCGGGCGGCGTGTACGGCTCGCCCACCGTCAACGGCAGCCGAGCCGGCGGGCTGATCGCGGCGTGCTGGGCCACCATGATGTTCGTGGGCAGACAGAGATACATTCAGATGACGAAGGAGGTGGTGGACACGGCGAGATACATCGAAAATGAATTACGCAAGATAGACGGCATCTTCATATTCGGCAAGCCCGCCACCACCGTCATCGCGTTCGGCTCCCAGCGGTTCGACATCTTCCAGCTGGCCGACCTGCTGCACAAGCGCGGCTGGGCGCTCAACGCGCTGCAGTTCCCGTCCGGGGTGCACCTGTGCGTGACGCACGCGCACACGTGCGACGGCGTGGCGGCGCGCTTCGTGGCGGACGTGGCGGCCGCCGCGCGCACCTGCCTCACCGACGCCGCGGCGCCAGTGCACGGCAAGATGGCGATATACGGCGTGGCGCAGGAGATATCGGACCGCAGCCTCGTGTCCGACATCACGAAGCACTTCATAGACTCCATGTACTACCTGCCCAAGGGCGACCCGCAGTAG
Protein
MSERPQPLRAINKFFDGKEPWQIVSMTASSLLAIVWVHSLYNARESVTVRLRKQFFRWLRQVPMVRKKIEQKMAEIKKDFHTDVEKRLAGVTIRRELPEQGLDPEQISQEVRDHLNLGAYDWKNGAVSGAVYHLCEDISRVACEAYTLTAYTNPLHSDVFPGINKMEAEIVQMAINLFNGDEECCGTVTTGGTESIIMACKAFRDLAYSKGISNPQIIVPSTAHSAFDKAAQYLGLFVVTIPVDPVTLCVDVDKVSDAIGRRTCLIVGSAPNYPYGTMDDIGALSALAARAGVPLHVDACLGGLVAGFMPAAGHPVPPFDFRLPGVASISADTHKYGYAPKGTSVILYRRAEYRHCQYTVTTEWPGGVYGSPTVNGSRAGGLIAACWATMMFVGRQRYIQMTKEVVDTARYIENELRKIDGIFIFGKPATTVIAFGSQRFDIFQLADLLHKRGWALNALQFPSGVHLCVTHAHTCDGVAARFVADVAAAARTCLTDAAAPVHGKMAIYGVAQEISDRSLVSDITKHFIDSMYYLPKGDPQ
Summary
Description
Cleaves phosphorylated sphingoid bases (PSBs), such as sphingosine-1-phosphate, into fatty aldehydes and phosphoethanolamine. Sphingolipid catabolism is required for normal development including viability, reproduction and muscle development.
Catalytic Activity
sphinganine 1-phosphate = hexadecanal + phosphoethanolamine
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the group II decarboxylase family.
Belongs to the CRISP family.
Belongs to the group II decarboxylase family. Sphingosine-1-phosphate lyase subfamily.
Belongs to the CRISP family.
Belongs to the group II decarboxylase family. Sphingosine-1-phosphate lyase subfamily.
Keywords
Complete proteome
Developmental protein
Endoplasmic reticulum
Lipid metabolism
Lyase
Membrane
Pyridoxal phosphate
Reference proteome
Signal-anchor
Sphingolipid metabolism
Transmembrane
Transmembrane helix
Feature
chain Sphingosine-1-phosphate lyase
Uniprot
A0A2W1BTK8
A0A194QEK6
A0A194QKC2
A0A2H1VXE7
D6W8W6
A0A146L8Z5
+ More
A0A1S4F3W7 A0A069DVF1 Q17G38 V5G2K7 A0A182GG03 A0A224XMN8 A0A195CRL9 A0A023F8H5 A0A034W6K0 X2D8H1 A0A0K8VL60 A0A0K8VUA2 A0A0A1X605 B3MD62 T1I8Q6 A0A1I8N556 A0A0P4VQY5 R4G5W3 A0A1W4W7L3 A0A1L8EHM1 U5EV11 A0A2M4CYU3 A0A087Z7V1 Q9V7Y2 A0A1L8EHT5 F4WLC4 B4HMH6 B4QIK3 T1P9B9 B4P8H4 A0A195FYI1 A0A2M3Z547 A0A2M4BJF9 A0A2M4BJH6 A0A2M4AIQ9 A0A2M4AHP3 A0A2A3E436 A0A158NZF1 A0A1L8EAP5 B0XFI0 A0A088AET0 A0A182FPV8 A0A1Q3FCN8 A0A1L8EAZ3 W8AGS9 A0A1B0B4E6 A0A0M4EIM4 A0A1A9YMW8 A0A067R790 A0A1Y1NA95 B3NP20 A0A1A9UGZ6 A0A2P8XBA2 A0A1L8E2I0 A0A151X3N5 J9JT04 A0A1I8P6T2 A0A1A9ZI41 A0A1W4VDG6 A0A1L8E2Z6 A0A182JU78 B4N5T0 Q290S2 A0A084VLC4 A0A182RZK1 B4J547 D3TNM0 A0A182Y3T6 A0A182VZM6 A0A1A9WWT6 A0A3B0J331 K7IMX0 B4LKG7 A0A182IPS7 A0A182Q4E5 A0A182M8F7 B4KQG4 A0A2H8TKP9 A0A182T279 A0A0M8ZT55 E2BL08 A0A182PSI3 Q7PY31 A0A182HLP2 A0A182WST9 A0A182VKI3 A0A2S2P5E8 A0A182LA33 A0A1B6H4D6 A0A182UED7 A0A154P1Y3 A0A182NRT2 J3JXX2
A0A1S4F3W7 A0A069DVF1 Q17G38 V5G2K7 A0A182GG03 A0A224XMN8 A0A195CRL9 A0A023F8H5 A0A034W6K0 X2D8H1 A0A0K8VL60 A0A0K8VUA2 A0A0A1X605 B3MD62 T1I8Q6 A0A1I8N556 A0A0P4VQY5 R4G5W3 A0A1W4W7L3 A0A1L8EHM1 U5EV11 A0A2M4CYU3 A0A087Z7V1 Q9V7Y2 A0A1L8EHT5 F4WLC4 B4HMH6 B4QIK3 T1P9B9 B4P8H4 A0A195FYI1 A0A2M3Z547 A0A2M4BJF9 A0A2M4BJH6 A0A2M4AIQ9 A0A2M4AHP3 A0A2A3E436 A0A158NZF1 A0A1L8EAP5 B0XFI0 A0A088AET0 A0A182FPV8 A0A1Q3FCN8 A0A1L8EAZ3 W8AGS9 A0A1B0B4E6 A0A0M4EIM4 A0A1A9YMW8 A0A067R790 A0A1Y1NA95 B3NP20 A0A1A9UGZ6 A0A2P8XBA2 A0A1L8E2I0 A0A151X3N5 J9JT04 A0A1I8P6T2 A0A1A9ZI41 A0A1W4VDG6 A0A1L8E2Z6 A0A182JU78 B4N5T0 Q290S2 A0A084VLC4 A0A182RZK1 B4J547 D3TNM0 A0A182Y3T6 A0A182VZM6 A0A1A9WWT6 A0A3B0J331 K7IMX0 B4LKG7 A0A182IPS7 A0A182Q4E5 A0A182M8F7 B4KQG4 A0A2H8TKP9 A0A182T279 A0A0M8ZT55 E2BL08 A0A182PSI3 Q7PY31 A0A182HLP2 A0A182WST9 A0A182VKI3 A0A2S2P5E8 A0A182LA33 A0A1B6H4D6 A0A182UED7 A0A154P1Y3 A0A182NRT2 J3JXX2
EC Number
4.1.2.27
Pubmed
28756777
26354079
18362917
19820115
26823975
26334808
+ More
17510324 26483478 25474469 25348373 25830018 17994087 18057021 25315136 27129103 20920257 12702658 10731132 12537572 12537569 28165339 21719571 22936249 17550304 21347285 24495485 24845553 28004739 29403074 15632085 23185243 24438588 20353571 25244985 20075255 20798317 12364791 14747013 17210077 20966253 22516182
17510324 26483478 25474469 25348373 25830018 17994087 18057021 25315136 27129103 20920257 12702658 10731132 12537572 12537569 28165339 21719571 22936249 17550304 21347285 24495485 24845553 28004739 29403074 15632085 23185243 24438588 20353571 25244985 20075255 20798317 12364791 14747013 17210077 20966253 22516182
EMBL
KZ149969
PZC76110.1
KQ459185
KPJ03380.1
KQ461198
KPJ05982.1
+ More
ODYU01004706 SOQ44874.1 KQ971312 EEZ98407.1 GDHC01014420 JAQ04209.1 GBGD01001098 JAC87791.1 CH477266 EAT45547.1 GALX01004216 JAB64250.1 JXUM01060987 KQ562129 KXJ76631.1 GFTR01007023 JAW09403.1 KQ977349 KYN03388.1 GBBI01001090 JAC17622.1 GAKP01009017 GAKP01009015 JAC49937.1 KC763805 AHJ81369.1 GDHF01012687 JAI39627.1 GDHF01009876 JAI42438.1 GBXI01014119 GBXI01007548 JAD00173.1 JAD06744.1 CH902619 EDV37395.1 KPU76786.1 ACPB03003517 GDKW01001757 JAI54838.1 GAHY01000170 JAA77340.1 GFDG01000645 JAV18154.1 GANO01002086 JAB57785.1 GGFL01006322 MBW70500.1 GGFL01006321 MBW70499.1 AJ297394 AE013599 AY052075 GFDG01000646 JAV18153.1 GL888207 EGI65087.1 CH480816 EDW48244.1 CM000362 CM002911 EDX07450.1 KMY94463.1 KA644503 AFP59132.1 CM000158 EDW92191.1 KQ981169 KYN45362.1 GGFM01002874 MBW23625.1 GGFJ01004068 MBW53209.1 GGFJ01004069 MBW53210.1 GGFK01007358 MBW40679.1 GGFK01006973 MBW40294.1 KZ288426 PBC25919.1 ADTU01004559 GFDG01003008 JAV15791.1 DS232930 EDS26818.1 GFDL01009787 JAV25258.1 GFDG01003007 JAV15792.1 GAMC01018970 JAB87585.1 JXJN01008282 CP012524 ALC40980.1 KK852898 KDR14164.1 GEZM01012147 GEZM01012146 JAV93146.1 CH954179 EDV55659.1 KQS62379.1 PYGN01004758 PSN29272.1 GFDF01001131 JAV12953.1 KQ982558 KYQ55001.1 ABLF02016317 GFDF01001132 JAV12952.1 CH964154 EDW79719.1 CM000071 EAL25290.1 KRT02047.1 ATLV01014444 KE524972 KFB38768.1 CH916367 EDW00673.1 EZ423022 ADD19298.1 OUUW01000001 SPP73603.1 CH940648 EDW60688.1 KRF79545.1 AXCN02001148 AXCM01000026 CH933808 EDW10304.1 GFXV01002427 MBW14232.1 KQ435890 KOX69456.1 GL448858 EFN83629.1 AAAB01008987 EAA00871.5 APCN01000909 GGMR01012016 MBY24635.1 GECZ01000210 JAS69559.1 KQ434803 KZC05959.1 BT128094 AEE63055.1
ODYU01004706 SOQ44874.1 KQ971312 EEZ98407.1 GDHC01014420 JAQ04209.1 GBGD01001098 JAC87791.1 CH477266 EAT45547.1 GALX01004216 JAB64250.1 JXUM01060987 KQ562129 KXJ76631.1 GFTR01007023 JAW09403.1 KQ977349 KYN03388.1 GBBI01001090 JAC17622.1 GAKP01009017 GAKP01009015 JAC49937.1 KC763805 AHJ81369.1 GDHF01012687 JAI39627.1 GDHF01009876 JAI42438.1 GBXI01014119 GBXI01007548 JAD00173.1 JAD06744.1 CH902619 EDV37395.1 KPU76786.1 ACPB03003517 GDKW01001757 JAI54838.1 GAHY01000170 JAA77340.1 GFDG01000645 JAV18154.1 GANO01002086 JAB57785.1 GGFL01006322 MBW70500.1 GGFL01006321 MBW70499.1 AJ297394 AE013599 AY052075 GFDG01000646 JAV18153.1 GL888207 EGI65087.1 CH480816 EDW48244.1 CM000362 CM002911 EDX07450.1 KMY94463.1 KA644503 AFP59132.1 CM000158 EDW92191.1 KQ981169 KYN45362.1 GGFM01002874 MBW23625.1 GGFJ01004068 MBW53209.1 GGFJ01004069 MBW53210.1 GGFK01007358 MBW40679.1 GGFK01006973 MBW40294.1 KZ288426 PBC25919.1 ADTU01004559 GFDG01003008 JAV15791.1 DS232930 EDS26818.1 GFDL01009787 JAV25258.1 GFDG01003007 JAV15792.1 GAMC01018970 JAB87585.1 JXJN01008282 CP012524 ALC40980.1 KK852898 KDR14164.1 GEZM01012147 GEZM01012146 JAV93146.1 CH954179 EDV55659.1 KQS62379.1 PYGN01004758 PSN29272.1 GFDF01001131 JAV12953.1 KQ982558 KYQ55001.1 ABLF02016317 GFDF01001132 JAV12952.1 CH964154 EDW79719.1 CM000071 EAL25290.1 KRT02047.1 ATLV01014444 KE524972 KFB38768.1 CH916367 EDW00673.1 EZ423022 ADD19298.1 OUUW01000001 SPP73603.1 CH940648 EDW60688.1 KRF79545.1 AXCN02001148 AXCM01000026 CH933808 EDW10304.1 GFXV01002427 MBW14232.1 KQ435890 KOX69456.1 GL448858 EFN83629.1 AAAB01008987 EAA00871.5 APCN01000909 GGMR01012016 MBY24635.1 GECZ01000210 JAS69559.1 KQ434803 KZC05959.1 BT128094 AEE63055.1
Proteomes
UP000053268
UP000053240
UP000007266
UP000008820
UP000069940
UP000249989
+ More
UP000078542 UP000007801 UP000015103 UP000095301 UP000192223 UP000000803 UP000007755 UP000001292 UP000000304 UP000002282 UP000078541 UP000242457 UP000005205 UP000002320 UP000005203 UP000069272 UP000092460 UP000092553 UP000092443 UP000027135 UP000008711 UP000078200 UP000245037 UP000075809 UP000007819 UP000095300 UP000092445 UP000192221 UP000075881 UP000007798 UP000001819 UP000030765 UP000075900 UP000001070 UP000076408 UP000075920 UP000091820 UP000268350 UP000002358 UP000008792 UP000075880 UP000075886 UP000075883 UP000009192 UP000075901 UP000053105 UP000008237 UP000075885 UP000007062 UP000075840 UP000076407 UP000075903 UP000075882 UP000075902 UP000076502 UP000075884
UP000078542 UP000007801 UP000015103 UP000095301 UP000192223 UP000000803 UP000007755 UP000001292 UP000000304 UP000002282 UP000078541 UP000242457 UP000005205 UP000002320 UP000005203 UP000069272 UP000092460 UP000092553 UP000092443 UP000027135 UP000008711 UP000078200 UP000245037 UP000075809 UP000007819 UP000095300 UP000092445 UP000192221 UP000075881 UP000007798 UP000001819 UP000030765 UP000075900 UP000001070 UP000076408 UP000075920 UP000091820 UP000268350 UP000002358 UP000008792 UP000075880 UP000075886 UP000075883 UP000009192 UP000075901 UP000053105 UP000008237 UP000075885 UP000007062 UP000075840 UP000076407 UP000075903 UP000075882 UP000075902 UP000076502 UP000075884
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BTK8
A0A194QEK6
A0A194QKC2
A0A2H1VXE7
D6W8W6
A0A146L8Z5
+ More
A0A1S4F3W7 A0A069DVF1 Q17G38 V5G2K7 A0A182GG03 A0A224XMN8 A0A195CRL9 A0A023F8H5 A0A034W6K0 X2D8H1 A0A0K8VL60 A0A0K8VUA2 A0A0A1X605 B3MD62 T1I8Q6 A0A1I8N556 A0A0P4VQY5 R4G5W3 A0A1W4W7L3 A0A1L8EHM1 U5EV11 A0A2M4CYU3 A0A087Z7V1 Q9V7Y2 A0A1L8EHT5 F4WLC4 B4HMH6 B4QIK3 T1P9B9 B4P8H4 A0A195FYI1 A0A2M3Z547 A0A2M4BJF9 A0A2M4BJH6 A0A2M4AIQ9 A0A2M4AHP3 A0A2A3E436 A0A158NZF1 A0A1L8EAP5 B0XFI0 A0A088AET0 A0A182FPV8 A0A1Q3FCN8 A0A1L8EAZ3 W8AGS9 A0A1B0B4E6 A0A0M4EIM4 A0A1A9YMW8 A0A067R790 A0A1Y1NA95 B3NP20 A0A1A9UGZ6 A0A2P8XBA2 A0A1L8E2I0 A0A151X3N5 J9JT04 A0A1I8P6T2 A0A1A9ZI41 A0A1W4VDG6 A0A1L8E2Z6 A0A182JU78 B4N5T0 Q290S2 A0A084VLC4 A0A182RZK1 B4J547 D3TNM0 A0A182Y3T6 A0A182VZM6 A0A1A9WWT6 A0A3B0J331 K7IMX0 B4LKG7 A0A182IPS7 A0A182Q4E5 A0A182M8F7 B4KQG4 A0A2H8TKP9 A0A182T279 A0A0M8ZT55 E2BL08 A0A182PSI3 Q7PY31 A0A182HLP2 A0A182WST9 A0A182VKI3 A0A2S2P5E8 A0A182LA33 A0A1B6H4D6 A0A182UED7 A0A154P1Y3 A0A182NRT2 J3JXX2
A0A1S4F3W7 A0A069DVF1 Q17G38 V5G2K7 A0A182GG03 A0A224XMN8 A0A195CRL9 A0A023F8H5 A0A034W6K0 X2D8H1 A0A0K8VL60 A0A0K8VUA2 A0A0A1X605 B3MD62 T1I8Q6 A0A1I8N556 A0A0P4VQY5 R4G5W3 A0A1W4W7L3 A0A1L8EHM1 U5EV11 A0A2M4CYU3 A0A087Z7V1 Q9V7Y2 A0A1L8EHT5 F4WLC4 B4HMH6 B4QIK3 T1P9B9 B4P8H4 A0A195FYI1 A0A2M3Z547 A0A2M4BJF9 A0A2M4BJH6 A0A2M4AIQ9 A0A2M4AHP3 A0A2A3E436 A0A158NZF1 A0A1L8EAP5 B0XFI0 A0A088AET0 A0A182FPV8 A0A1Q3FCN8 A0A1L8EAZ3 W8AGS9 A0A1B0B4E6 A0A0M4EIM4 A0A1A9YMW8 A0A067R790 A0A1Y1NA95 B3NP20 A0A1A9UGZ6 A0A2P8XBA2 A0A1L8E2I0 A0A151X3N5 J9JT04 A0A1I8P6T2 A0A1A9ZI41 A0A1W4VDG6 A0A1L8E2Z6 A0A182JU78 B4N5T0 Q290S2 A0A084VLC4 A0A182RZK1 B4J547 D3TNM0 A0A182Y3T6 A0A182VZM6 A0A1A9WWT6 A0A3B0J331 K7IMX0 B4LKG7 A0A182IPS7 A0A182Q4E5 A0A182M8F7 B4KQG4 A0A2H8TKP9 A0A182T279 A0A0M8ZT55 E2BL08 A0A182PSI3 Q7PY31 A0A182HLP2 A0A182WST9 A0A182VKI3 A0A2S2P5E8 A0A182LA33 A0A1B6H4D6 A0A182UED7 A0A154P1Y3 A0A182NRT2 J3JXX2
PDB
4Q6R
E-value=7.97794e-121,
Score=1111
Ontologies
PATHWAY
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
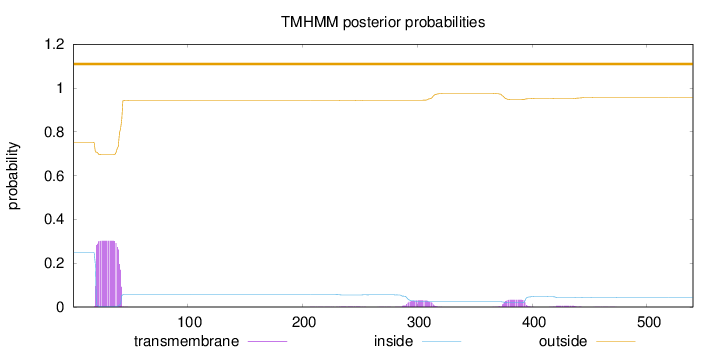
Length:
540
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.95812999999998
Exp number, first 60 AAs:
6.48741
Total prob of N-in:
0.24922
outside
1 - 540
Population Genetic Test Statistics
Pi
187.228813
Theta
155.45265
Tajima's D
0.45979
CLR
0.354445
CSRT
0.509374531273436
Interpretation
Uncertain
