Gene
KWMTBOMO04880 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008020
Annotation
PREDICTED:_ras_suppressor_protein_1_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.489 Nuclear Reliability : 1.944
Sequence
CDS
ATGAGTCATCAACCCCCAGTATCTTGTATTCCAAACACAGGCAAGATGTCGAAAGCTAAAAAAGTTATTGAAGAAGCCAAAGAAATAAATAATCCGGAAATCGATTTAGTCGACAAAGGAATATCTAGTCTCGAAGAGATTCCAGGCTTATTTTCTCTGGAGAACATCACTCGTCTGTCCCTAAGCCACAACAAAATCTCTGTAGTGCCTGCTGCACTAGCCAACTTGAGCAACTTAGAAATTCTAAACTTGGCCAATAACAACATTCAAGAGCTTCCAGTCAGCTTATCATCACTACCCAAGTTAAGGATCCTCAATGTTTCATTAAACAAACTGTATAACTTGCCAAGAGGCTTTGGTTCCTTCCCTGTGCTAGAGATATTGGATCTAACATACAATAATTTGAATGAAAAAGTATTACCTGGGAACTTCTTTATTATGGACAGTCTCCGTGCTTTGTATTTGGGTGACAATGATTTTGAATTTCTACCACCTGAAATTGGAAATTTGAAGAATCTACAAATTTTGTCGATGCGCGAGAACGACCTGATCGAAGTTCCCCGGGAGCTGGGGCAGCTGGCGCGCCTCCGCGAGCTGCACCTGCAGGGGAACCGCCTCGTCGTGCTGCCGCCGGAGATCGGAACATTGGACCTCGCCAGCAACAAGTCAGTGCTGAGGCTGGAAGGTAACTTCTGGGTTCCGCCCATCGAAGACCAGCTCAAGCTGGGGCCGTCCCACGTGCTGGACTATCTGCGCTCAGAGACCTACAAAGTCCTGTACAGCCGCCACATGTCGGCCAAGCCGCCGCCGCCGCCGCAGACCGTCGACAAGAGCAAGAAGGCGAGTCGCGCCGCCTAA
Protein
MSHQPPVSCIPNTGKMSKAKKVIEEAKEINNPEIDLVDKGISSLEEIPGLFSLENITRLSLSHNKISVVPAALANLSNLEILNLANNNIQELPVSLSSLPKLRILNVSLNKLYNLPRGFGSFPVLEILDLTYNNLNEKVLPGNFFIMDSLRALYLGDNDFEFLPPEIGNLKNLQILSMRENDLIEVPRELGQLARLRELHLQGNRLVVLPPEIGTLDLASNKSVLRLEGNFWVPPIEDQLKLGPSHVLDYLRSETYKVLYSRHMSAKPPPPPQTVDKSKKASRAA
Summary
Uniprot
A0A2D2AK62
A0A2H1VVK9
A0A194QK84
H9JES3
A0A2D2AK49
A0A2W1BM21
+ More
A0A212EJT5 S4PCD8 A0A194QIV1 A0A2P8XTF6 A0A067RQW7 A0A2J7QDG5 A0A0C9R2M8 A0A026X515 A0A1Y1JRH2 A0A154PU41 A0A195CJW6 A0A151XBI8 A0A151I274 A0A195F0Z4 A0A158NG42 A0A088ATP7 A0A1B6KGW7 A0A3L8DNK2 E2B4K0 A0A195E7D0 E2A8L4 A0A0N0U4P1 A0A310SJ72 A0A1B6K6W1 A0A0T6ATY5 A0A0L7QLN4 A0A1B6GI68 T1JL02 K7J9I2 B3NAU1 B4IEP5 B4Q542 A0A2S2QWF4 Q9V428 A0A1W4UV66 A0A2H8TM86 A0A1B6DSG8 D6WCX1 A0A2S2N7C2 B4P3N5 B3MV41 B4N0C2 B5DHC6 B4GKG0 A0A3B0JN67 J9JT46 A0A0L0CJP8 R4G5F4 F4WTP1 A0A0A1X947 A0A1A9X2A5 A0A1B0BFK3 A0A1I8N6Y0 Q7JVW9 T1HGL5 A0A0K8V6U2 B4M9R3 A0A293N649 A0A224YSP9 A0A131YJF6 A0A131XHK9 A0A1I8P1K3 E9H6T9 L7M660 A0A090X8E1 B7P3B9 B4KIQ9 A0A1E1X840 A0A0P5W5S3 A0A1A9ZHV0 D3TMM8 A0A1L8DLZ6 A0A1A9V158 G3MJE9 W8B5G1 A0A0A9YWP7 A0A1S4F2J0 A0A1L8EDU9 A0A0P4VSB0 A0A1Q3FCX3 T1PEQ3 A0A0M4ER08 U4UDI7 E9IRF2 B0XCG1 A0A023EN75 A0A023EM33 A0A210Q4P4 A0A182GBM1 A0A1L8EBT7 A0A1L8EBI0 A0A1L8EBC7 A0A023EM30
A0A212EJT5 S4PCD8 A0A194QIV1 A0A2P8XTF6 A0A067RQW7 A0A2J7QDG5 A0A0C9R2M8 A0A026X515 A0A1Y1JRH2 A0A154PU41 A0A195CJW6 A0A151XBI8 A0A151I274 A0A195F0Z4 A0A158NG42 A0A088ATP7 A0A1B6KGW7 A0A3L8DNK2 E2B4K0 A0A195E7D0 E2A8L4 A0A0N0U4P1 A0A310SJ72 A0A1B6K6W1 A0A0T6ATY5 A0A0L7QLN4 A0A1B6GI68 T1JL02 K7J9I2 B3NAU1 B4IEP5 B4Q542 A0A2S2QWF4 Q9V428 A0A1W4UV66 A0A2H8TM86 A0A1B6DSG8 D6WCX1 A0A2S2N7C2 B4P3N5 B3MV41 B4N0C2 B5DHC6 B4GKG0 A0A3B0JN67 J9JT46 A0A0L0CJP8 R4G5F4 F4WTP1 A0A0A1X947 A0A1A9X2A5 A0A1B0BFK3 A0A1I8N6Y0 Q7JVW9 T1HGL5 A0A0K8V6U2 B4M9R3 A0A293N649 A0A224YSP9 A0A131YJF6 A0A131XHK9 A0A1I8P1K3 E9H6T9 L7M660 A0A090X8E1 B7P3B9 B4KIQ9 A0A1E1X840 A0A0P5W5S3 A0A1A9ZHV0 D3TMM8 A0A1L8DLZ6 A0A1A9V158 G3MJE9 W8B5G1 A0A0A9YWP7 A0A1S4F2J0 A0A1L8EDU9 A0A0P4VSB0 A0A1Q3FCX3 T1PEQ3 A0A0M4ER08 U4UDI7 E9IRF2 B0XCG1 A0A023EN75 A0A023EM33 A0A210Q4P4 A0A182GBM1 A0A1L8EBT7 A0A1L8EBI0 A0A1L8EBC7 A0A023EM30
Pubmed
26354079
19121390
28756777
22118469
23622113
29403074
+ More
24845553 24508170 28004739 21347285 30249741 20798317 20075255 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 17550304 15632085 26108605 21719571 25830018 25315136 28797301 26830274 28049606 21292972 25576852 25970599 28503490 20353571 22216098 24495485 25401762 27129103 23537049 21282665 24945155 28812685 26483478
24845553 24508170 28004739 21347285 30249741 20798317 20075255 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 17550304 15632085 26108605 21719571 25830018 25315136 28797301 26830274 28049606 21292972 25576852 25970599 28503490 20353571 22216098 24495485 25401762 27129103 23537049 21282665 24945155 28812685 26483478
EMBL
KY073283
ATQ37759.1
ODYU01004706
SOQ44875.1
KQ461198
KPJ05983.1
+ More
BABH01023375 KY073282 ATQ37758.1 KZ149969 PZC76109.1 AGBW02014406 OWR41730.1 GAIX01004231 JAA88329.1 KQ459185 KPJ03381.1 PYGN01001375 PSN35308.1 KK852476 KDR23045.1 NEVH01015820 PNF26634.1 GBYB01010399 JAG80166.1 KK107020 EZA62519.1 GEZM01102787 JAV51792.1 KQ435180 KZC14964.1 KQ977721 KYN00364.1 KQ982335 KYQ57658.1 KQ976580 KYM79954.1 KQ981864 KYN34143.1 ADTU01014960 GEBQ01029265 JAT10712.1 QOIP01000006 RLU21802.1 GL445574 EFN89353.1 KQ979568 KYN21001.1 GL437652 EFN70201.1 KQ435812 KOX72844.1 KQ760559 OAD59795.1 GECU01000514 JAT07193.1 LJIG01022880 KRT78254.1 KQ414915 KOC59459.1 GECZ01007649 JAS62120.1 JH431739 CH954177 EDV58655.1 CH480831 EDW46072.1 CM000361 CM002910 EDX04948.1 KMY90135.1 GGMS01012259 MBY81462.1 AE014134 BT031285 BT088923 AAF53320.2 ABY20526.1 ACT35687.1 AHN54416.1 AHN54417.1 GFXV01003295 MBW15100.1 GEDC01008703 JAS28595.1 KQ971311 EEZ99069.1 GGMR01000410 MBY13029.1 CM000157 EDW88336.2 CH902624 EDV33106.2 CH963920 EDW77535.2 CH379058 EDY69701.1 CH479184 EDW37126.1 OUUW01000006 SPP82313.1 ABLF02035210 ABLF02035217 JRES01000305 KNC32471.1 GAHY01000016 JAA77494.1 GL888341 EGI62462.1 GBXI01006861 GBXI01005428 GBXI01001850 JAD07431.1 JAD08864.1 JAD12442.1 JXJN01013525 AY118581 AAM49950.1 ACPB03018566 GDHF01031961 GDHF01030563 GDHF01029388 GDHF01025297 GDHF01025088 GDHF01022447 GDHF01019048 GDHF01017713 GDHF01007622 GDHF01006894 JAI20353.1 JAI21751.1 JAI22926.1 JAI27017.1 JAI27226.1 JAI29867.1 JAI33266.1 JAI34601.1 JAI44692.1 JAI45420.1 CH940654 EDW57939.2 GFWV01022941 MAA47668.1 GFPF01009492 MAA20638.1 GEDV01010361 JAP78196.1 GEFH01002973 JAP65608.1 GL732598 EFX72571.1 GACK01005677 JAA59357.1 GBIH01001968 JAC92742.1 ABJB010060055 ABJB010346723 ABJB010477874 ABJB010681524 DS627523 EEC01091.1 CH933807 EDW12415.1 GFAC01003751 JAT95437.1 GDIQ01227669 GDIP01090867 LRGB01002912 JAK24056.1 JAM12848.1 KZS05522.1 CCAG010013018 EZ422680 ADD18956.1 GFDF01006605 JAV07479.1 JO842000 AEO33617.1 GAMC01014202 GAMC01014201 GAMC01014200 JAB92353.1 GBHO01009610 GBHO01009609 JAG33994.1 JAG33995.1 GFDG01001975 JAV16824.1 GDKW01001781 JAI54814.1 GFDL01009619 JAV25426.1 KA646403 AFP61032.1 CP012523 ALC39525.1 KB632309 ERL92039.1 GL765108 EFZ16851.1 DS232689 EDS44791.1 GAPW01003258 JAC10340.1 GAPW01003257 JAC10341.1 NEDP02005024 OWF43695.1 JXUM01052821 KQ561750 KXJ77640.1 GFDG01002776 JAV16023.1 GFDG01002775 JAV16024.1 GFDG01002893 JAV15906.1 GAPW01003256 JAC10342.1
BABH01023375 KY073282 ATQ37758.1 KZ149969 PZC76109.1 AGBW02014406 OWR41730.1 GAIX01004231 JAA88329.1 KQ459185 KPJ03381.1 PYGN01001375 PSN35308.1 KK852476 KDR23045.1 NEVH01015820 PNF26634.1 GBYB01010399 JAG80166.1 KK107020 EZA62519.1 GEZM01102787 JAV51792.1 KQ435180 KZC14964.1 KQ977721 KYN00364.1 KQ982335 KYQ57658.1 KQ976580 KYM79954.1 KQ981864 KYN34143.1 ADTU01014960 GEBQ01029265 JAT10712.1 QOIP01000006 RLU21802.1 GL445574 EFN89353.1 KQ979568 KYN21001.1 GL437652 EFN70201.1 KQ435812 KOX72844.1 KQ760559 OAD59795.1 GECU01000514 JAT07193.1 LJIG01022880 KRT78254.1 KQ414915 KOC59459.1 GECZ01007649 JAS62120.1 JH431739 CH954177 EDV58655.1 CH480831 EDW46072.1 CM000361 CM002910 EDX04948.1 KMY90135.1 GGMS01012259 MBY81462.1 AE014134 BT031285 BT088923 AAF53320.2 ABY20526.1 ACT35687.1 AHN54416.1 AHN54417.1 GFXV01003295 MBW15100.1 GEDC01008703 JAS28595.1 KQ971311 EEZ99069.1 GGMR01000410 MBY13029.1 CM000157 EDW88336.2 CH902624 EDV33106.2 CH963920 EDW77535.2 CH379058 EDY69701.1 CH479184 EDW37126.1 OUUW01000006 SPP82313.1 ABLF02035210 ABLF02035217 JRES01000305 KNC32471.1 GAHY01000016 JAA77494.1 GL888341 EGI62462.1 GBXI01006861 GBXI01005428 GBXI01001850 JAD07431.1 JAD08864.1 JAD12442.1 JXJN01013525 AY118581 AAM49950.1 ACPB03018566 GDHF01031961 GDHF01030563 GDHF01029388 GDHF01025297 GDHF01025088 GDHF01022447 GDHF01019048 GDHF01017713 GDHF01007622 GDHF01006894 JAI20353.1 JAI21751.1 JAI22926.1 JAI27017.1 JAI27226.1 JAI29867.1 JAI33266.1 JAI34601.1 JAI44692.1 JAI45420.1 CH940654 EDW57939.2 GFWV01022941 MAA47668.1 GFPF01009492 MAA20638.1 GEDV01010361 JAP78196.1 GEFH01002973 JAP65608.1 GL732598 EFX72571.1 GACK01005677 JAA59357.1 GBIH01001968 JAC92742.1 ABJB010060055 ABJB010346723 ABJB010477874 ABJB010681524 DS627523 EEC01091.1 CH933807 EDW12415.1 GFAC01003751 JAT95437.1 GDIQ01227669 GDIP01090867 LRGB01002912 JAK24056.1 JAM12848.1 KZS05522.1 CCAG010013018 EZ422680 ADD18956.1 GFDF01006605 JAV07479.1 JO842000 AEO33617.1 GAMC01014202 GAMC01014201 GAMC01014200 JAB92353.1 GBHO01009610 GBHO01009609 JAG33994.1 JAG33995.1 GFDG01001975 JAV16824.1 GDKW01001781 JAI54814.1 GFDL01009619 JAV25426.1 KA646403 AFP61032.1 CP012523 ALC39525.1 KB632309 ERL92039.1 GL765108 EFZ16851.1 DS232689 EDS44791.1 GAPW01003258 JAC10340.1 GAPW01003257 JAC10341.1 NEDP02005024 OWF43695.1 JXUM01052821 KQ561750 KXJ77640.1 GFDG01002776 JAV16023.1 GFDG01002775 JAV16024.1 GFDG01002893 JAV15906.1 GAPW01003256 JAC10342.1
Proteomes
UP000053240
UP000005204
UP000007151
UP000053268
UP000245037
UP000027135
+ More
UP000235965 UP000053097 UP000076502 UP000078542 UP000075809 UP000078540 UP000078541 UP000005205 UP000005203 UP000279307 UP000008237 UP000078492 UP000000311 UP000053105 UP000053825 UP000002358 UP000008711 UP000001292 UP000000304 UP000000803 UP000192221 UP000007266 UP000002282 UP000007801 UP000007798 UP000001819 UP000008744 UP000268350 UP000007819 UP000037069 UP000007755 UP000091820 UP000092460 UP000095301 UP000015103 UP000008792 UP000095300 UP000000305 UP000001555 UP000009192 UP000076858 UP000092445 UP000092444 UP000078200 UP000092553 UP000030742 UP000002320 UP000242188 UP000069940 UP000249989
UP000235965 UP000053097 UP000076502 UP000078542 UP000075809 UP000078540 UP000078541 UP000005205 UP000005203 UP000279307 UP000008237 UP000078492 UP000000311 UP000053105 UP000053825 UP000002358 UP000008711 UP000001292 UP000000304 UP000000803 UP000192221 UP000007266 UP000002282 UP000007801 UP000007798 UP000001819 UP000008744 UP000268350 UP000007819 UP000037069 UP000007755 UP000091820 UP000092460 UP000095301 UP000015103 UP000008792 UP000095300 UP000000305 UP000001555 UP000009192 UP000076858 UP000092445 UP000092444 UP000078200 UP000092553 UP000030742 UP000002320 UP000242188 UP000069940 UP000249989
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2D2AK62
A0A2H1VVK9
A0A194QK84
H9JES3
A0A2D2AK49
A0A2W1BM21
+ More
A0A212EJT5 S4PCD8 A0A194QIV1 A0A2P8XTF6 A0A067RQW7 A0A2J7QDG5 A0A0C9R2M8 A0A026X515 A0A1Y1JRH2 A0A154PU41 A0A195CJW6 A0A151XBI8 A0A151I274 A0A195F0Z4 A0A158NG42 A0A088ATP7 A0A1B6KGW7 A0A3L8DNK2 E2B4K0 A0A195E7D0 E2A8L4 A0A0N0U4P1 A0A310SJ72 A0A1B6K6W1 A0A0T6ATY5 A0A0L7QLN4 A0A1B6GI68 T1JL02 K7J9I2 B3NAU1 B4IEP5 B4Q542 A0A2S2QWF4 Q9V428 A0A1W4UV66 A0A2H8TM86 A0A1B6DSG8 D6WCX1 A0A2S2N7C2 B4P3N5 B3MV41 B4N0C2 B5DHC6 B4GKG0 A0A3B0JN67 J9JT46 A0A0L0CJP8 R4G5F4 F4WTP1 A0A0A1X947 A0A1A9X2A5 A0A1B0BFK3 A0A1I8N6Y0 Q7JVW9 T1HGL5 A0A0K8V6U2 B4M9R3 A0A293N649 A0A224YSP9 A0A131YJF6 A0A131XHK9 A0A1I8P1K3 E9H6T9 L7M660 A0A090X8E1 B7P3B9 B4KIQ9 A0A1E1X840 A0A0P5W5S3 A0A1A9ZHV0 D3TMM8 A0A1L8DLZ6 A0A1A9V158 G3MJE9 W8B5G1 A0A0A9YWP7 A0A1S4F2J0 A0A1L8EDU9 A0A0P4VSB0 A0A1Q3FCX3 T1PEQ3 A0A0M4ER08 U4UDI7 E9IRF2 B0XCG1 A0A023EN75 A0A023EM33 A0A210Q4P4 A0A182GBM1 A0A1L8EBT7 A0A1L8EBI0 A0A1L8EBC7 A0A023EM30
A0A212EJT5 S4PCD8 A0A194QIV1 A0A2P8XTF6 A0A067RQW7 A0A2J7QDG5 A0A0C9R2M8 A0A026X515 A0A1Y1JRH2 A0A154PU41 A0A195CJW6 A0A151XBI8 A0A151I274 A0A195F0Z4 A0A158NG42 A0A088ATP7 A0A1B6KGW7 A0A3L8DNK2 E2B4K0 A0A195E7D0 E2A8L4 A0A0N0U4P1 A0A310SJ72 A0A1B6K6W1 A0A0T6ATY5 A0A0L7QLN4 A0A1B6GI68 T1JL02 K7J9I2 B3NAU1 B4IEP5 B4Q542 A0A2S2QWF4 Q9V428 A0A1W4UV66 A0A2H8TM86 A0A1B6DSG8 D6WCX1 A0A2S2N7C2 B4P3N5 B3MV41 B4N0C2 B5DHC6 B4GKG0 A0A3B0JN67 J9JT46 A0A0L0CJP8 R4G5F4 F4WTP1 A0A0A1X947 A0A1A9X2A5 A0A1B0BFK3 A0A1I8N6Y0 Q7JVW9 T1HGL5 A0A0K8V6U2 B4M9R3 A0A293N649 A0A224YSP9 A0A131YJF6 A0A131XHK9 A0A1I8P1K3 E9H6T9 L7M660 A0A090X8E1 B7P3B9 B4KIQ9 A0A1E1X840 A0A0P5W5S3 A0A1A9ZHV0 D3TMM8 A0A1L8DLZ6 A0A1A9V158 G3MJE9 W8B5G1 A0A0A9YWP7 A0A1S4F2J0 A0A1L8EDU9 A0A0P4VSB0 A0A1Q3FCX3 T1PEQ3 A0A0M4ER08 U4UDI7 E9IRF2 B0XCG1 A0A023EN75 A0A023EM33 A0A210Q4P4 A0A182GBM1 A0A1L8EBT7 A0A1L8EBI0 A0A1L8EBC7 A0A023EM30
PDB
4U08
E-value=2.78516e-16,
Score=207
Ontologies
GO
PANTHER
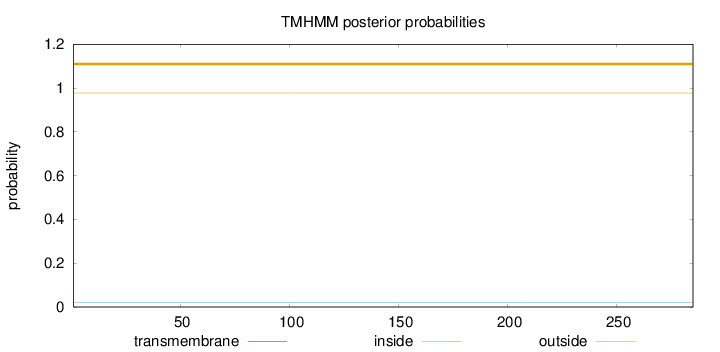
Topology
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00254
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.02220
outside
1 - 285
Population Genetic Test Statistics
Pi
32.694034
Theta
41.100034
Tajima's D
-1.341235
CLR
429.008326
CSRT
0.0788960551972401
Interpretation
Uncertain
