Gene
KWMTBOMO04879
Pre Gene Modal
BGIBMGA008052
Annotation
PREDICTED:_nuclear_envelope_integral_membrane_protein_1a_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.924
Sequence
CDS
ATGATTAGTGAATTAAGGATATCGGTGTTGTTAATGCTTCTATTATCAGAAGCGTCTTCTTTACCTTACGACATTCATTGGTTAGGCGGACCGACCACAATTGAAAGGGAAATACCATCTTCGAAAAAGGCTATCGACATCTACTGTTATACTGGTACAACAAAAAGTTTATTTGCACTCTGGCAAACTGTGAAATTTCAAATAAGAATTAAAAATGATGAATTTCGTCAATACCTTGGCTCAAATCCAGAAGATGTTTATAAAGAGTATACAAAAGATAGCTACGGTTGGTCTGTTGTTAATCCGTTCATTAAGAAATCACAGAAAACTGTTTCCATCAACTTGTTTGAGCCAACATGCATGGCCATTAACACTAAAGAAAGACATAGTATTGACCTGCTGGTGCAAAGGGTGGATCTATGGCGCGTCATACTAATGGCTGCTGGCATAGCACTCATCTTCTCTTCAAAGTCTCTGAGCGGTAACCCCGTGTTCTTCTACTTGTGTGGGATCTTCATTGGCGTCTTTGCATCATTTATGGTTTTAATGTATTACATCAGCAAAATGCTGCCTAAGCGCACATTAACTTATGGCCTCCTCATTGGTGGCTGGACTGTTGGTGTGTATCTATTCCAACAAATGTGGGAGAACATTCAAACATTAGCTGTGGATCACCGGAATTATGTGATCTGGTACACGTTGGCAGTCAGCTTCATCAGTTTCTTAGTTTGCTACAGGATAGGTCCACCGAGGAATCAACGAAGCAAGAACATTGTAATGTGGACAATACAGTGCGTCGGAGTGCTGATGATATTCTTTAGCAGCGAGTACCAGGAAGCTTCAGCGGCCATCATCATAGCTTCGATGATCATTAAGTATTTCCCGGAGTCGTGGGTATACAGAATACGAGGATATTGGCGCAGCAAGTTCCCCCCGGCGCGGCGCCTGCTCACCAGCGAGGAGTACTACGAGCAGGGAGCGCGCGAGACGCGGCGCGCGCTGGCGGCGCTGCGGGAGCACTGCGCCAGCCCCGACTGCGCGCAGTGGGCCGTCATGCTCAAACTTAACGACTCCAAGAGATTCGCAAGTTTCGTGGAGGGGAATTCGCACCTCAGCGACGACGAGATCCTGGACTACGAGTCCTACGCGTTCTCCCTGGACCGGAGCCGCAAGCCCAGCTCGCACTCCACCAGGAACCAGCTCGAGATATCTGACGACGACGACCTCACCGACAGCGACGACGACTCCGCGTAG
Protein
MISELRISVLLMLLLSEASSLPYDIHWLGGPTTIEREIPSSKKAIDIYCYTGTTKSLFALWQTVKFQIRIKNDEFRQYLGSNPEDVYKEYTKDSYGWSVVNPFIKKSQKTVSINLFEPTCMAINTKERHSIDLLVQRVDLWRVILMAAGIALIFSSKSLSGNPVFFYLCGIFIGVFASFMVLMYYISKMLPKRTLTYGLLIGGWTVGVYLFQQMWENIQTLAVDHRNYVIWYTLAVSFISFLVCYRIGPPRNQRSKNIVMWTIQCVGVLMIFFSSEYQEASAAIIIASMIIKYFPESWVYRIRGYWRSKFPPARRLLTSEEYYEQGARETRRALAALREHCASPDCAQWAVMLKLNDSKRFASFVEGNSHLSDDEILDYESYAFSLDRSRKPSSHSTRNQLEISDDDDLTDSDDDSA
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
H9JEV5
A0A2H1VVU7
A0A2W1BME9
A0A194QD44
A0A194QKR1
A0A212EJR3
+ More
D6WCX2 A0A1W7R4V9 U5EQP6 A0A067RJR7 B0WSD1 A0A182GE98 A0A1B0D4C7 A0A0L0C0G0 A0A2J7QF68 A0A1Y1KIZ8 N6U565 U4UDI7 A0A1L8D9A1 Q16L76 A0A1S4G4N1 Q1DH09 A0A1I8Q0M6 A0A1Q3F4F4 A0A1Q3F5N7 A0A0L7KSX5 A0A336M6Z1 A0A1A9YDC7 A0A1B0B045 B4L8J9 A0A1I8MUB5 B4F668 B4M1Y0 A0A1A9VC14 B4F666 B4NES7 B4F667 A0A1B0A3I9 C0MM50 C0MM52 B4F677 B4PXV9 B4R605 B4F672 Q9VXD6 C0MM51 A0A1B0CA28 B3NVK3 A0A1W4VVM8 B4GV93 C0MM57 D1GYY0 W8BP99 B3MW98 D1GYX6 D1GYY1 D1GYY2 D1GYX9 D1GZ13 D1GYY5 A0A0K8UH58 A0A1B0A594 D1GYX8 A0A1B0G429 A0A0A1XHF4 D1GZ41 B4IEQ8 A0A3B0JYW3 B5DLI4 B4JLT1 A0A0A9WCK4 A0A0M3QZ18 T1HEX7 A0A1A9WII1 B5DLI2 A0A182RJY2 A0A1B6C6W0 A0A182TIX2 A0A182XNV1 A0A182Y0F7 A0A182I7F6 E0VB39 A0NGE1 A0A1A9X553 A0A1S4GZQ7 A0A182V0I1 W5JDD3 A0A182F1L8 A0A2M4BPR0 A0A1J1HJI0 A0A182JWV4 A0A182WG98 A0A026WDX8 A0A182LMN2 A0A182QJB5 A0A1Z5LGB7
D6WCX2 A0A1W7R4V9 U5EQP6 A0A067RJR7 B0WSD1 A0A182GE98 A0A1B0D4C7 A0A0L0C0G0 A0A2J7QF68 A0A1Y1KIZ8 N6U565 U4UDI7 A0A1L8D9A1 Q16L76 A0A1S4G4N1 Q1DH09 A0A1I8Q0M6 A0A1Q3F4F4 A0A1Q3F5N7 A0A0L7KSX5 A0A336M6Z1 A0A1A9YDC7 A0A1B0B045 B4L8J9 A0A1I8MUB5 B4F668 B4M1Y0 A0A1A9VC14 B4F666 B4NES7 B4F667 A0A1B0A3I9 C0MM50 C0MM52 B4F677 B4PXV9 B4R605 B4F672 Q9VXD6 C0MM51 A0A1B0CA28 B3NVK3 A0A1W4VVM8 B4GV93 C0MM57 D1GYY0 W8BP99 B3MW98 D1GYX6 D1GYY1 D1GYY2 D1GYX9 D1GZ13 D1GYY5 A0A0K8UH58 A0A1B0A594 D1GYX8 A0A1B0G429 A0A0A1XHF4 D1GZ41 B4IEQ8 A0A3B0JYW3 B5DLI4 B4JLT1 A0A0A9WCK4 A0A0M3QZ18 T1HEX7 A0A1A9WII1 B5DLI2 A0A182RJY2 A0A1B6C6W0 A0A182TIX2 A0A182XNV1 A0A182Y0F7 A0A182I7F6 E0VB39 A0NGE1 A0A1A9X553 A0A1S4GZQ7 A0A182V0I1 W5JDD3 A0A182F1L8 A0A2M4BPR0 A0A1J1HJI0 A0A182JWV4 A0A182WG98 A0A026WDX8 A0A182LMN2 A0A182QJB5 A0A1Z5LGB7
Pubmed
19121390
28756777
26354079
22118469
18362917
19820115
+ More
24845553 26483478 26108605 28004739 23537049 17510324 26227816 17994087 25315136 18477586 19126864 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 18057021 25830018 15632085 25401762 26823975 25244985 20566863 12364791 20920257 23761445 24508170 30249741 20966253 28528879
24845553 26483478 26108605 28004739 23537049 17510324 26227816 17994087 25315136 18477586 19126864 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 18057021 25830018 15632085 25401762 26823975 25244985 20566863 12364791 20920257 23761445 24508170 30249741 20966253 28528879
EMBL
BABH01023375
ODYU01004706
SOQ44876.1
KZ149969
PZC76108.1
KQ459185
+ More
KPJ03382.1 KQ461198 KPJ05984.1 AGBW02014406 OWR41729.1 KQ971311 EEZ98828.1 GEHC01001454 JAV46191.1 GANO01004162 JAB55709.1 KK852427 KDR24091.1 DS232068 EDS33769.1 JXUM01057463 KQ561956 KXJ77058.1 AJVK01011357 JRES01001072 KNC25746.1 NEVH01015302 PNF27193.1 GEZM01087327 JAV58767.1 APGK01042240 KB741002 ENN75781.1 KB632309 ERL92039.1 GFDF01011157 JAV02927.1 CH477918 EAT35075.1 CH899885 EAT32440.1 GFDL01012600 JAV22445.1 GFDL01012178 JAV22867.1 JTDY01006204 KOB66200.1 UFQS01000642 UFQT01000642 SSX05677.1 SSX26036.1 JXJN01006553 CH933815 EDW07974.1 AM999304 AM999305 AM999306 AM999307 AM999309 AM999310 AM999311 AM999312 FM246421 FM246425 FM246427 FM246430 CAQ53666.1 CAQ53667.1 CAQ53668.1 CAQ53669.1 CAQ53671.1 CAQ53672.1 CAQ53673.1 CAQ53674.1 CAR94347.1 CAR94351.1 CAR94353.1 CAR94356.1 CH940651 EDW65684.2 AM999302 CAQ53664.1 CH964239 EDW82246.2 AM999303 CAQ53665.1 FM246422 CAR94348.1 FM246424 FM246426 FM246428 CAR94350.1 CAR94352.1 CAR94354.1 AM999313 CAQ53675.1 CM000162 EDX01945.1 CM000366 EDX18109.1 AM999308 CAQ53670.1 AE014298 AY069627 AAF48638.1 AAL39772.1 FM246423 CAR94349.1 AJWK01003181 CH954180 EDV46395.1 CH479192 EDW26630.1 FM246429 CAR94355.1 FN546719 CBE67018.1 GAMC01003595 JAC02961.1 CH902625 EDV35243.2 KPU75494.1 FN546715 CBE67014.1 FN546716 FN546720 FN546723 CBE67015.1 CBE67019.1 CBE67022.1 FN546721 FN546722 CBE67020.1 CBE67021.1 FN546718 CBE67017.1 FN546752 CBE67051.1 FN546724 CBE67023.1 GDHF01026297 JAI26017.1 FN546717 CBE67016.1 CCAG010020285 GBXI01005555 GBXI01003876 JAD08737.1 JAD10416.1 FN546780 CBE67079.1 CH480832 EDW46162.1 OUUW01000003 SPP77921.1 CH379063 EDY72385.2 CH916371 EDV91692.1 GBHO01038072 GBHO01038070 GBHO01038069 GBHO01038068 GBHO01001057 GDHC01013866 JAG05532.1 JAG05534.1 JAG05535.1 JAG05536.1 JAG42547.1 JAQ04763.1 CP012528 ALC48653.1 ACPB03020693 ACPB03020694 EDY72383.2 GEDC01028066 JAS09232.1 APCN01003394 DS235021 EEB10595.1 AAAB01008984 EAU75915.2 ADMH02001861 ETN60824.1 GGFJ01005863 MBW55004.1 CVRI01000004 CRK87714.1 KK107293 QOIP01000005 EZA53234.1 RLU22573.1 AXCN02000739 GFJQ02000916 JAW06054.1
KPJ03382.1 KQ461198 KPJ05984.1 AGBW02014406 OWR41729.1 KQ971311 EEZ98828.1 GEHC01001454 JAV46191.1 GANO01004162 JAB55709.1 KK852427 KDR24091.1 DS232068 EDS33769.1 JXUM01057463 KQ561956 KXJ77058.1 AJVK01011357 JRES01001072 KNC25746.1 NEVH01015302 PNF27193.1 GEZM01087327 JAV58767.1 APGK01042240 KB741002 ENN75781.1 KB632309 ERL92039.1 GFDF01011157 JAV02927.1 CH477918 EAT35075.1 CH899885 EAT32440.1 GFDL01012600 JAV22445.1 GFDL01012178 JAV22867.1 JTDY01006204 KOB66200.1 UFQS01000642 UFQT01000642 SSX05677.1 SSX26036.1 JXJN01006553 CH933815 EDW07974.1 AM999304 AM999305 AM999306 AM999307 AM999309 AM999310 AM999311 AM999312 FM246421 FM246425 FM246427 FM246430 CAQ53666.1 CAQ53667.1 CAQ53668.1 CAQ53669.1 CAQ53671.1 CAQ53672.1 CAQ53673.1 CAQ53674.1 CAR94347.1 CAR94351.1 CAR94353.1 CAR94356.1 CH940651 EDW65684.2 AM999302 CAQ53664.1 CH964239 EDW82246.2 AM999303 CAQ53665.1 FM246422 CAR94348.1 FM246424 FM246426 FM246428 CAR94350.1 CAR94352.1 CAR94354.1 AM999313 CAQ53675.1 CM000162 EDX01945.1 CM000366 EDX18109.1 AM999308 CAQ53670.1 AE014298 AY069627 AAF48638.1 AAL39772.1 FM246423 CAR94349.1 AJWK01003181 CH954180 EDV46395.1 CH479192 EDW26630.1 FM246429 CAR94355.1 FN546719 CBE67018.1 GAMC01003595 JAC02961.1 CH902625 EDV35243.2 KPU75494.1 FN546715 CBE67014.1 FN546716 FN546720 FN546723 CBE67015.1 CBE67019.1 CBE67022.1 FN546721 FN546722 CBE67020.1 CBE67021.1 FN546718 CBE67017.1 FN546752 CBE67051.1 FN546724 CBE67023.1 GDHF01026297 JAI26017.1 FN546717 CBE67016.1 CCAG010020285 GBXI01005555 GBXI01003876 JAD08737.1 JAD10416.1 FN546780 CBE67079.1 CH480832 EDW46162.1 OUUW01000003 SPP77921.1 CH379063 EDY72385.2 CH916371 EDV91692.1 GBHO01038072 GBHO01038070 GBHO01038069 GBHO01038068 GBHO01001057 GDHC01013866 JAG05532.1 JAG05534.1 JAG05535.1 JAG05536.1 JAG42547.1 JAQ04763.1 CP012528 ALC48653.1 ACPB03020693 ACPB03020694 EDY72383.2 GEDC01028066 JAS09232.1 APCN01003394 DS235021 EEB10595.1 AAAB01008984 EAU75915.2 ADMH02001861 ETN60824.1 GGFJ01005863 MBW55004.1 CVRI01000004 CRK87714.1 KK107293 QOIP01000005 EZA53234.1 RLU22573.1 AXCN02000739 GFJQ02000916 JAW06054.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000007266
UP000027135
+ More
UP000002320 UP000069940 UP000249989 UP000092462 UP000037069 UP000235965 UP000019118 UP000030742 UP000008820 UP000095300 UP000037510 UP000092443 UP000092460 UP000009192 UP000095301 UP000008792 UP000078200 UP000007798 UP000092445 UP000002282 UP000000304 UP000000803 UP000092461 UP000008711 UP000192221 UP000008744 UP000007801 UP000092444 UP000001292 UP000268350 UP000001819 UP000001070 UP000092553 UP000015103 UP000091820 UP000075900 UP000075902 UP000076407 UP000076408 UP000075840 UP000009046 UP000007062 UP000075903 UP000000673 UP000069272 UP000183832 UP000075881 UP000075920 UP000053097 UP000279307 UP000075882 UP000075886
UP000002320 UP000069940 UP000249989 UP000092462 UP000037069 UP000235965 UP000019118 UP000030742 UP000008820 UP000095300 UP000037510 UP000092443 UP000092460 UP000009192 UP000095301 UP000008792 UP000078200 UP000007798 UP000092445 UP000002282 UP000000304 UP000000803 UP000092461 UP000008711 UP000192221 UP000008744 UP000007801 UP000092444 UP000001292 UP000268350 UP000001819 UP000001070 UP000092553 UP000015103 UP000091820 UP000075900 UP000075902 UP000076407 UP000076408 UP000075840 UP000009046 UP000007062 UP000075903 UP000000673 UP000069272 UP000183832 UP000075881 UP000075920 UP000053097 UP000279307 UP000075882 UP000075886
Pfam
Interpro
IPR036259
MFS_trans_sf
+ More
IPR019358 NEMP_fam
IPR032412 Myosin-VI_CBD
IPR036114 MYSc_Myo6
IPR000048 IQ_motif_EF-hand-BS
IPR027417 P-loop_NTPase
IPR020846 MFS_dom
IPR001609 Myosin_head_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR005475 Transketolase-like_Pyr-bd
IPR001017 DH_E1
IPR031717 KGD_C
IPR011603 2oxoglutarate_DH_E1
IPR042179 KGD_C_sf
IPR029061 THDP-binding
IPR019358 NEMP_fam
IPR032412 Myosin-VI_CBD
IPR036114 MYSc_Myo6
IPR000048 IQ_motif_EF-hand-BS
IPR027417 P-loop_NTPase
IPR020846 MFS_dom
IPR001609 Myosin_head_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR005475 Transketolase-like_Pyr-bd
IPR001017 DH_E1
IPR031717 KGD_C
IPR011603 2oxoglutarate_DH_E1
IPR042179 KGD_C_sf
IPR029061 THDP-binding
Gene 3D
ProteinModelPortal
H9JEV5
A0A2H1VVU7
A0A2W1BME9
A0A194QD44
A0A194QKR1
A0A212EJR3
+ More
D6WCX2 A0A1W7R4V9 U5EQP6 A0A067RJR7 B0WSD1 A0A182GE98 A0A1B0D4C7 A0A0L0C0G0 A0A2J7QF68 A0A1Y1KIZ8 N6U565 U4UDI7 A0A1L8D9A1 Q16L76 A0A1S4G4N1 Q1DH09 A0A1I8Q0M6 A0A1Q3F4F4 A0A1Q3F5N7 A0A0L7KSX5 A0A336M6Z1 A0A1A9YDC7 A0A1B0B045 B4L8J9 A0A1I8MUB5 B4F668 B4M1Y0 A0A1A9VC14 B4F666 B4NES7 B4F667 A0A1B0A3I9 C0MM50 C0MM52 B4F677 B4PXV9 B4R605 B4F672 Q9VXD6 C0MM51 A0A1B0CA28 B3NVK3 A0A1W4VVM8 B4GV93 C0MM57 D1GYY0 W8BP99 B3MW98 D1GYX6 D1GYY1 D1GYY2 D1GYX9 D1GZ13 D1GYY5 A0A0K8UH58 A0A1B0A594 D1GYX8 A0A1B0G429 A0A0A1XHF4 D1GZ41 B4IEQ8 A0A3B0JYW3 B5DLI4 B4JLT1 A0A0A9WCK4 A0A0M3QZ18 T1HEX7 A0A1A9WII1 B5DLI2 A0A182RJY2 A0A1B6C6W0 A0A182TIX2 A0A182XNV1 A0A182Y0F7 A0A182I7F6 E0VB39 A0NGE1 A0A1A9X553 A0A1S4GZQ7 A0A182V0I1 W5JDD3 A0A182F1L8 A0A2M4BPR0 A0A1J1HJI0 A0A182JWV4 A0A182WG98 A0A026WDX8 A0A182LMN2 A0A182QJB5 A0A1Z5LGB7
D6WCX2 A0A1W7R4V9 U5EQP6 A0A067RJR7 B0WSD1 A0A182GE98 A0A1B0D4C7 A0A0L0C0G0 A0A2J7QF68 A0A1Y1KIZ8 N6U565 U4UDI7 A0A1L8D9A1 Q16L76 A0A1S4G4N1 Q1DH09 A0A1I8Q0M6 A0A1Q3F4F4 A0A1Q3F5N7 A0A0L7KSX5 A0A336M6Z1 A0A1A9YDC7 A0A1B0B045 B4L8J9 A0A1I8MUB5 B4F668 B4M1Y0 A0A1A9VC14 B4F666 B4NES7 B4F667 A0A1B0A3I9 C0MM50 C0MM52 B4F677 B4PXV9 B4R605 B4F672 Q9VXD6 C0MM51 A0A1B0CA28 B3NVK3 A0A1W4VVM8 B4GV93 C0MM57 D1GYY0 W8BP99 B3MW98 D1GYX6 D1GYY1 D1GYY2 D1GYX9 D1GZ13 D1GYY5 A0A0K8UH58 A0A1B0A594 D1GYX8 A0A1B0G429 A0A0A1XHF4 D1GZ41 B4IEQ8 A0A3B0JYW3 B5DLI4 B4JLT1 A0A0A9WCK4 A0A0M3QZ18 T1HEX7 A0A1A9WII1 B5DLI2 A0A182RJY2 A0A1B6C6W0 A0A182TIX2 A0A182XNV1 A0A182Y0F7 A0A182I7F6 E0VB39 A0NGE1 A0A1A9X553 A0A1S4GZQ7 A0A182V0I1 W5JDD3 A0A182F1L8 A0A2M4BPR0 A0A1J1HJI0 A0A182JWV4 A0A182WG98 A0A026WDX8 A0A182LMN2 A0A182QJB5 A0A1Z5LGB7
Ontologies
GO
Topology
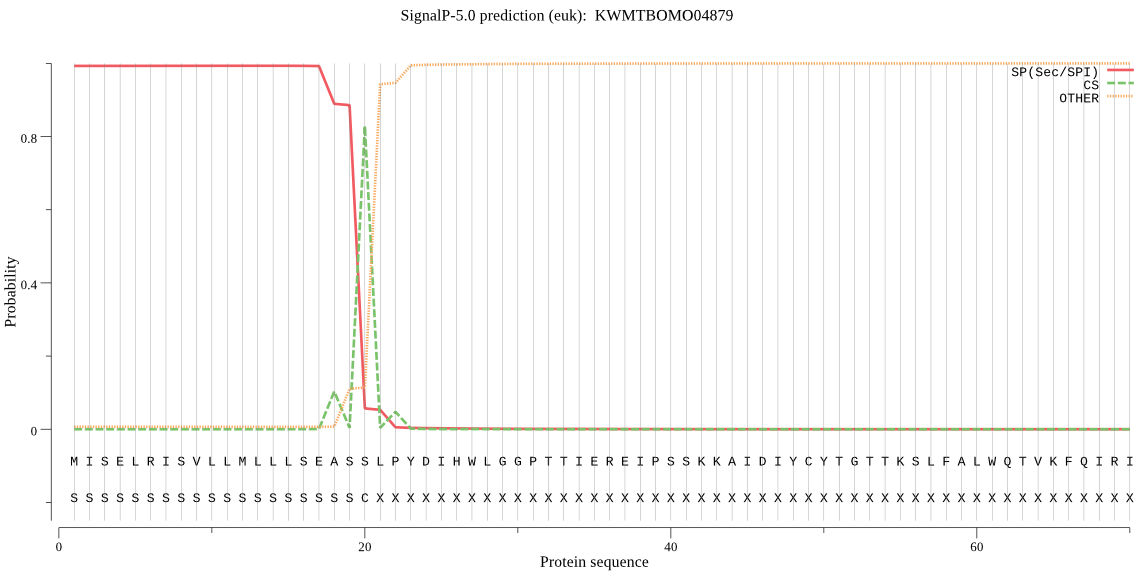
SignalP
Position: 1 - 20,
Likelihood: 0.993121
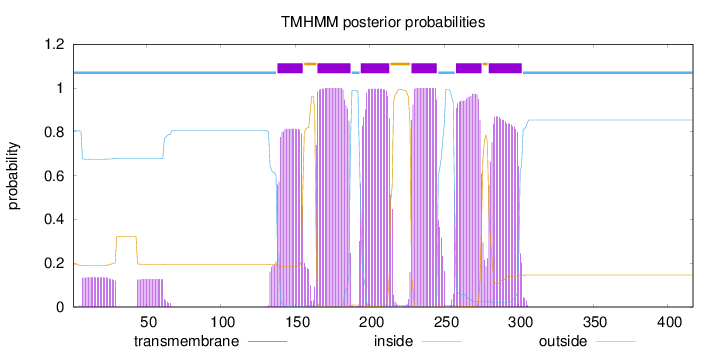
Length:
417
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
119.14917
Exp number, first 60 AAs:
5.24147
Total prob of N-in:
0.80698
inside
1 - 137
TMhelix
138 - 155
outside
156 - 164
TMhelix
165 - 187
inside
188 - 193
TMhelix
194 - 213
outside
214 - 227
TMhelix
228 - 245
inside
246 - 257
TMhelix
258 - 275
outside
276 - 279
TMhelix
280 - 302
inside
303 - 417
Population Genetic Test Statistics
Pi
182.990631
Theta
170.912534
Tajima's D
0.172406
CLR
0.770079
CSRT
0.416429178541073
Interpretation
Uncertain
