Pre Gene Modal
BGIBMGA008021
Annotation
death_related_ced-3/Nedd2-like_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.497 Nuclear Reliability : 2.186
Sequence
CDS
ATGCAATCTTCGAGTCCTGGACCTGTTATTGATCTTCGTGCTGATGTTAACCGCCTTGTGTTGCACAGCAACCTTATAAACTTGGAAGTGATTATCGACATTGAACACGAACTAAGGAAAGAATCTGGTGACCTTATTTCCCTTATGTTTCTCTTGTACGATGATCCAGACATTGCATTAGTAAAATTGACAGCTTATCAGCAATCTAGTAATGCATATCATGAGCATTCCTTGCTTCAAGATTGGCTTTTAAAGGCAAAATGTAAGCTGACCTGGAAACATGAATTGGTCGAAGGATTGTTAATTTGCAAAATGAATAATTTGATTAGAAAGATGGGTTTTGATTTAAACAGCTTAAAAACATACTACCAAACAGACAATTGCATAATGAGCATACATATAAATCCTGTAAAAAAGTTACTTTACAGAATATGCGAGGAAATGAATGCCACGAATTTTTCGAAGTTCAAACGGTCGCTCTTGGCCTATGGTTTGGATTTTACAAAGCAAGACATTTGTGAAATTATCTTGCTTGAATTAATATCTAAAGAATTTATACAGACGAATTTTAGTAACAAATGTTTATCCGTTGCAAATACTTCTACTCAAAAAATGGCCGTGCGGGATTTCGTGAAAACAATTGAAAATTTAGAAGGGTTACAAATATTGGCAATGGAACTAAACGCTTTAGAGACGGAAGTGAATAAACATTTCGAAAAGGCTTCTAATTCATCTGAGGACGCCCCTATTTCGGAAGAAGCACATAAAGAAGACGATAAATTTGATAATGCTGAAATATTTGAAGATTCTTTGAATTTTGAAAATCTGACATTAGAAGATTTTCATGCGAGTTGTTAG
Protein
MQSSSPGPVIDLRADVNRLVLHSNLINLEVIIDIEHELRKESGDLISLMFLLYDDPDIALVKLTAYQQSSNAYHEHSLLQDWLLKAKCKLTWKHELVEGLLICKMNNLIRKMGFDLNSLKTYYQTDNCIMSIHINPVKKLLYRICEEMNATNFSKFKRSLLAYGLDFTKQDICEIILLELISKEFIQTNFSNKCLSVANTSTQKMAVRDFVKTIENLEGLQILAMELNALETEVNKHFEKASNSSEDAPISEEAHKEDDKFDNAEIFEDSLNFENLTLEDFHASC
Summary
Similarity
Belongs to the peptidase C14A family.
Uniprot
Pubmed
EMBL
BABH01023371
AB292816
BAF98475.1
BABH01008520
JTDY01000408
KOB77332.1
+ More
HQ328981 AEK20838.1 NWSH01000001 PCG81126.1 HQ328982 AEK20839.1 HQ328983 AEK20840.1 JX040310 AFO64608.1 AGBW02009100 OWR51688.1 KQ461198 KPJ06283.1 JQ768052 AFJ04536.1 KU668855 AMR71144.1 KQ458793 KPJ05070.1 MH200927 AXS59137.1 HM234679 AEF30497.1 GDQN01001913 JAT89141.1 HQ328980 AEK20837.1 CH478203 EAT33580.1 DQ860099 ABI74776.1 JXUM01104218 KQ564868 KXJ71641.1
HQ328981 AEK20838.1 NWSH01000001 PCG81126.1 HQ328982 AEK20839.1 HQ328983 AEK20840.1 JX040310 AFO64608.1 AGBW02009100 OWR51688.1 KQ461198 KPJ06283.1 JQ768052 AFJ04536.1 KU668855 AMR71144.1 KQ458793 KPJ05070.1 MH200927 AXS59137.1 HM234679 AEF30497.1 GDQN01001913 JAT89141.1 HQ328980 AEK20837.1 CH478203 EAT33580.1 DQ860099 ABI74776.1 JXUM01104218 KQ564868 KXJ71641.1
Proteomes
Interpro
SUPFAM
SSF52129
SSF52129
ProteinModelPortal
Ontologies
PANTHER
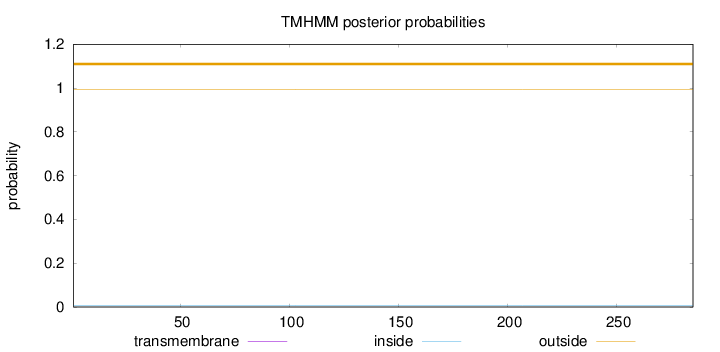
Topology
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000580000000000001
Exp number, first 60 AAs:
0.00033
Total prob of N-in:
0.00789
outside
1 - 285
Population Genetic Test Statistics
Pi
102.137143
Theta
116.60135
Tajima's D
-0.451704
CLR
0.678258
CSRT
0.250737463126844
Interpretation
Uncertain
