Gene
KWMTBOMO04877 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008051
Annotation
PREDICTED:_myosin_heavy_chain_95F_isoform_X2_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.372 Nuclear Reliability : 1.851
Sequence
CDS
ATGACGATCGACAAGCAGTTAGTATGGGTCCGGGAGCCGAACGAGGGCTTCGTCCTGGCGCGGCTCCACGAGCTCATGGGCGAGGAGGCGGACGTGGTGCCCCTGGACAACAAGTACCCGAGGAGGACTTGCAGCTTCGAGGATATATTCCCTGCTGGAGATCCCAATAAAGATGTCGACGACAACTGCGAGCTGATGTTCTTGAATGAAGCAACACTCCTGAATAATATCTTGACTCGTTACAAGAAGAACAAGATATACACGTATGTGGCGAATATTCTACTGGCGGTAAATCCTTACAAAGAAATAGCAGATCTGTATTCTTCGAACACGATAAAAATGTATCAAGGGAAATCTTTGGGACAACTTCCGCCTCATGTTTTTGCTATCGCCGACAAAGCGTTCAGAGACATGAAAGCGTTCAAGCAGTCGCAATCCATAATCGTGTCGGGCGAGTCCGGCGCCGGCAAGACGGAGTCCACCAAGTACATCCTCAAGTACCTGTGCGAGCTGTGGGCCAAGAGCGCGGGGCCCGTCGAACAGAAGCTACTAGACGCGAACCCTATCCTCGAAGCGTTCGGCAACGCGAAGACGACCCGCAACAACAACAGCTCGCGCTTCGGCAAGTTCATCGAGGTGCACTTCAGCGGCGCGTGCAGCGTCACCGGCGGACACATCTCGCACTACCTGCTCGAGCGCTCGCGCGTGTGTCGCCAGGCGCCCGCCGAGCGGAACTACCACGTGTTCTACCTGCTGTGTGCCGGCGCGCCGCCCCCCCTGCGCACCTACCTGCGCCTCACCAAGCCAGACGACTACTACTACCTCAAGAGCGGCTGCACCCAGTACTTCACTACGCCGGACTCGGAAAAGAAGATCAGCGCCGACCGCAAGAGTAAAGATCACCTCGCCAAGGGCGGGCTGCGGGACCCCATACTTGACGACTATGAAGATTTCCAGAGACTTAATCAGGCGCTGAGTCGCGCCGGCTTGTCGGAGGAGGAGAAGCGCGACGTGTTCGGGGCCGTGGCCGGCGCCCTGCACCTCGGGAACATCACGTTCGACGAGGAGCCCGGCGCGCGCGGGGGCTGCGCCGTGCACGCACACGCCGAGCCCGCGCTCGCCGCCGCCGCCGCCCTCTTCCACGTCCCCGCCGACATGCTGCGCCGCGCCCTCGTGTCCCGCCTCATGCAGAGCGCGCGCGCCGGCCACAAGGGCACCGCCATCATGGTCCCGCTGAAGCCGCACGAGGCGTGCAGCGCCCGCGACGCGCTGGCCAAGGCGGTGTACGGCCGCGTGTTCGAGCACGTGGTGCGCCGCGTCAACCGAGCCGTGCCAGCCGTCGCCGCGCCCTACTACGTCGGCGTGCTGGACATCGCCGGCTTCGAGTACTTCGAGATGAACAGCTTCGAGCAGTTCTGCATAAATTACTGCAACGAGAAACTGCAGCAGTTCTTCAACGAGCGCATTCTCAAGAACGAGCAGGAGCTGTACAAGCGGGAGGGACTCAACGTGCCCCACATACAGTACGTCGACAACCAGGACTGCATCGATTTGATCGAGAGCAAGAACCACGGCATATTCCACTACCTGGACGAGGAGTCCAAGCTGCCCAAGCCGGAGCCGTCCCACTTCACGGAGTGCGTGCACCGGGAGCTGGGCGCGCACAGCGAGCGCCTGCACGTGCCGCGCTCCTCCCGCCTCAAGGCGCACCGCGAGCTGCGGGACCACGAGGGCTTCCTGCTGCGACACTTCGCCGGCGCCGTCTGCTACAGCACGGGGCAGTTCATCGAGAAGAACAACGACGCCCTGCACGCGTCCCTGGGCGTGCTCATGCAGGAGTCGGACAGCGCCTTCGTGCGCGAGCTGTTCGAGGAGGCGGGCGCGGAGCCGGCGCGGGGCCGCCTCGCCTTCCAGTCCGTCGGCGCCAAGTTCCAGGCGCAGCTGGCGCAGCTCATGGACAAGCTCGGCAAGAACGGCACGAAGTTCGTCCGCTGCGTCAAGCCCAACGGCTCCATGCGGGGCGGCGCGGGCGAGGGCGGCCTGGTGCTGGCGCAGCTGCAGTGCAGCGGCACCGGCGCCGCGCTGCAGCTCATGGAGCACGGCTACCCCTCGCGCGCCCCCTTCCTGCACCTGCACCACACCTACGCGCCGCACCTGCCGCACCAGCTCGCCAGCCTGTCGCCCCGGACCTTCTGCGAGGCGATGGTGCACAGCCTGGGGCTGTCCGACAAGGACTTCAAGCTGGGCGTGAGCCGCGTGTTCTTCCGCCCCGGCAAGTACAGCGAGCTGGACGCGCTGCTGCGCTCCGAGCCGCACGAGCTGCAGGCCGTCGTCAAGAACGTGCTGACGTGGCTCGTCAGGTCGCGCTGGAGGAAGAGCATCTTCGCTGTGCTTTCCATTATCAAATTGAAGAATAAGATAGTGTACCGGCGCTCGTGCCTGGTGCTGATCCAGAAGTGCGTGCGCGGGCACGTGGTGCGGCGCCGCCACCGCCCGCGCCTGGCCGCGCTGCGCCAGCTCGCCGCGCTGCGACACAACCTGGTCGACATGCGGCGCGCGCTCGCCAGCCTGCGCCCCGCCCCGCCCGCCGCGCACCCCGCGCTCGACGCGCTGGCCCGCCGGCTGGACGACGCCGCCCGCGCCGTCAAGGACAACGAGCACATGTCCCGCGAGCAGCTGGACAGCATGCAGGCCACACTGTCCAAAGAAGTCGAAGTACAGATGAATGCGATACAGAAGGCCGTAGTCGAACAGAAGAACAGGGAGGAACAAGAGAGACTTCGCAAGATCGAAGCGGAGATCCGAGCCGCCGAGGAAGCGAAGCGCATCGAGCAGGAGAAGATCCGACAGGAGGAGGAGAACAGGAGGCTGAAAGCGGAGATGGAGGCGCGGCGCAAGGAAGAGGAGCGTCTGCGTCACCAGCAGCCGGCCGACGTCGCCGCTGACGTCATCGACCCGCTGCAGCAGGACCGCACCGACCGGGACCGGCTGGAGCAGGAGCGCCGGGACCACGAGATGGCGCTGCGCCTCGCCCGGGAGACCAACGGACACGTGGAGGGCTCCCCGCCGCAACTGCGCAGGTCGGAGCGCGTGCGGCTGCAGCAGGCCGCGCACGGCAAGCAGAAGCACGACCTCTCCAAGTGGAAGTACTCCGAGCTCAGAGACACCATCAACACCTCCTGCGACATCGAGCTGCTCGAGGCCTGCAGACACGAGTTCCACCGCCGCCTGAAGGTGTACCACGCGTGGAAGGCGAAGAACGCGCGCAAGTCCACGCTGCACGAGCAGGAGCGGGCGCCGCAGTCCGTCATGGACGCCGCGAAGAGTCCTCGCGTACTGCAGAAGGGCGATATCCTAGGCGCCCGCCACCGCTACTTCCGCATACCGTTCGTCCGCGGAGGGGCGGGGGGCGACGGCGGCACGCGCGGCTGGTGGTACGCGCACTTCGACGGGCAGTACGTGGCGCGGCAGATGGAGCTGCACCCCGACAAGACGCCCGTGCTGCTGCGCGCCGGCGCCGACGACATGCAGATGTGCGAGCTCAGCCTCGACGAGACCGGCCTCACCCGCAAGCGCGGCGCCGAGATACTGCCGCACGAGTTCGAGCGCCAGTGGACCGCGCACGGCGGGCCCGCCTACGCGCCGCCGCCCCGCTAG
Protein
MTIDKQLVWVREPNEGFVLARLHELMGEEADVVPLDNKYPRRTCSFEDIFPAGDPNKDVDDNCELMFLNEATLLNNILTRYKKNKIYTYVANILLAVNPYKEIADLYSSNTIKMYQGKSLGQLPPHVFAIADKAFRDMKAFKQSQSIIVSGESGAGKTESTKYILKYLCELWAKSAGPVEQKLLDANPILEAFGNAKTTRNNNSSRFGKFIEVHFSGACSVTGGHISHYLLERSRVCRQAPAERNYHVFYLLCAGAPPPLRTYLRLTKPDDYYYLKSGCTQYFTTPDSEKKISADRKSKDHLAKGGLRDPILDDYEDFQRLNQALSRAGLSEEEKRDVFGAVAGALHLGNITFDEEPGARGGCAVHAHAEPALAAAAALFHVPADMLRRALVSRLMQSARAGHKGTAIMVPLKPHEACSARDALAKAVYGRVFEHVVRRVNRAVPAVAAPYYVGVLDIAGFEYFEMNSFEQFCINYCNEKLQQFFNERILKNEQELYKREGLNVPHIQYVDNQDCIDLIESKNHGIFHYLDEESKLPKPEPSHFTECVHRELGAHSERLHVPRSSRLKAHRELRDHEGFLLRHFAGAVCYSTGQFIEKNNDALHASLGVLMQESDSAFVRELFEEAGAEPARGRLAFQSVGAKFQAQLAQLMDKLGKNGTKFVRCVKPNGSMRGGAGEGGLVLAQLQCSGTGAALQLMEHGYPSRAPFLHLHHTYAPHLPHQLASLSPRTFCEAMVHSLGLSDKDFKLGVSRVFFRPGKYSELDALLRSEPHELQAVVKNVLTWLVRSRWRKSIFAVLSIIKLKNKIVYRRSCLVLIQKCVRGHVVRRRHRPRLAALRQLAALRHNLVDMRRALASLRPAPPAAHPALDALARRLDDAARAVKDNEHMSREQLDSMQATLSKEVEVQMNAIQKAVVEQKNREEQERLRKIEAEIRAAEEAKRIEQEKIRQEEENRRLKAEMEARRKEEERLRHQQPADVAADVIDPLQQDRTDRDRLEQERRDHEMALRLARETNGHVEGSPPQLRRSERVRLQQAAHGKQKHDLSKWKYSELRDTINTSCDIELLEACRHEFHRRLKVYHAWKAKNARKSTLHEQERAPQSVMDAAKSPRVLQKGDILGARHRYFRIPFVRGGAGGDGGTRGWWYAHFDGQYVARQMELHPDKTPVLLRAGADDMQMCELSLDETGLTRKRGAEILPHEFERQWTAHGGPAYAPPPR
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
A0A212EJV6
A0A194QD44
A0A1S4F6Z8
A0A182H7Y9
A0A1Q3FZD7
A0A154PFV1
+ More
A0A1S4F759 Q17DE9 U5EW23 A0A1Q3FZH6 A0A0J7KVX9 A0A2A3E163 A0A195EHC0 A0A151WQY6 E2C7U5 A0A1A9W6M3 A0A034V7N4 A0A1J1J5A6 A0A182FK19 A0A034V9W9 A0A084VJE1 B4G4F6 B5DYV4 A0A026WT79 A0A2M4BA63 A0A224XD56 A0A069DXG8 I5ANW8 W5JLL4 I5ANW9 A0A3B0K493 A0A3B0KGM1 A0A3B0K9Q5 A0A146L0Q7 A0A146LJJ8 E0VU00 N6UJR9 A0A2M4A5V3 E9HEA9 A0A0P5NPA2 A0A0P6DBF0 A0A0P5D452 Q7QE85 A0A0P6HLF0 A0A0P5SU58 A0A182MI83 A0A2H8TF30 A0A0P5D5E0 J9JYL7 A0A2S2QQP9 A0A0P6GI71 A0A0P6F7L5 A0A131Y2K9 V5HPH1 A0A0N8E1X3 A0A2M3Z2Q6 V5I894 A0A2M4A5D9 A0A0K8VWF7 A0A0K8UQX8 A0A0K8VHC1 A0A1E1XM31 A0A023PNK0 A0A2T7P4G3 A0A1B6DN64 R7U9G6 A0A1B6CMV6 A0A2C9JLB9 A0A3R7N3Q3 A0A267EZ74 A0A267EY54 A0A0P5Q1N5 A0A1Q3G4Q3 A0A088AJQ9 A0A0K8VTR8 A0A0K8WF13 A0A0K8WMD2 A0A0K8VFU5 A0A310S7H3 A0A0M8ZUX6 A0A195CBB8 A0A2M3Z322 D6W8W8 A0A158NHU4 A0A195AXB1 A0A158NHU5 A0A0C9R0T8 A0A0C9QE67 A0A1B6GYM3 A0A1B6FAM9 A0A1B6M2P4 A0A1B6LJ37 A0A2J7PRL7 A0A2J7PRL6 A0A2J7PRN3 A0A1Y1MI87 A0A0P5A5E9 A0A182VBR0 A0A182I9K4 A0A067QY55
A0A1S4F759 Q17DE9 U5EW23 A0A1Q3FZH6 A0A0J7KVX9 A0A2A3E163 A0A195EHC0 A0A151WQY6 E2C7U5 A0A1A9W6M3 A0A034V7N4 A0A1J1J5A6 A0A182FK19 A0A034V9W9 A0A084VJE1 B4G4F6 B5DYV4 A0A026WT79 A0A2M4BA63 A0A224XD56 A0A069DXG8 I5ANW8 W5JLL4 I5ANW9 A0A3B0K493 A0A3B0KGM1 A0A3B0K9Q5 A0A146L0Q7 A0A146LJJ8 E0VU00 N6UJR9 A0A2M4A5V3 E9HEA9 A0A0P5NPA2 A0A0P6DBF0 A0A0P5D452 Q7QE85 A0A0P6HLF0 A0A0P5SU58 A0A182MI83 A0A2H8TF30 A0A0P5D5E0 J9JYL7 A0A2S2QQP9 A0A0P6GI71 A0A0P6F7L5 A0A131Y2K9 V5HPH1 A0A0N8E1X3 A0A2M3Z2Q6 V5I894 A0A2M4A5D9 A0A0K8VWF7 A0A0K8UQX8 A0A0K8VHC1 A0A1E1XM31 A0A023PNK0 A0A2T7P4G3 A0A1B6DN64 R7U9G6 A0A1B6CMV6 A0A2C9JLB9 A0A3R7N3Q3 A0A267EZ74 A0A267EY54 A0A0P5Q1N5 A0A1Q3G4Q3 A0A088AJQ9 A0A0K8VTR8 A0A0K8WF13 A0A0K8WMD2 A0A0K8VFU5 A0A310S7H3 A0A0M8ZUX6 A0A195CBB8 A0A2M3Z322 D6W8W8 A0A158NHU4 A0A195AXB1 A0A158NHU5 A0A0C9R0T8 A0A0C9QE67 A0A1B6GYM3 A0A1B6FAM9 A0A1B6M2P4 A0A1B6LJ37 A0A2J7PRL7 A0A2J7PRL6 A0A2J7PRN3 A0A1Y1MI87 A0A0P5A5E9 A0A182VBR0 A0A182I9K4 A0A067QY55
Pubmed
EMBL
AGBW02014406
OWR41728.1
KQ459185
KPJ03382.1
JXUM01028664
JXUM01028665
+ More
KQ560828 KXJ80718.1 GFDL01002114 JAV32931.1 KQ434896 KZC10725.1 CH477296 EAT44388.1 GANO01001616 JAB58255.1 GFDL01002115 JAV32930.1 LBMM01002703 KMQ94458.1 KZ288466 PBC25438.1 KQ978957 KYN27267.1 KQ982813 KYQ50276.1 GL453466 EFN75980.1 GAKP01019656 GAKP01019654 JAC39296.1 CVRI01000069 CRL06988.1 GAKP01019655 GAKP01019653 JAC39299.1 ATLV01013545 KE524867 KFB38085.1 CH479179 EDW24504.1 CM000070 EDY68029.3 KK107109 EZA59148.1 GGFJ01000775 MBW49916.1 GFTR01008718 JAW07708.1 GBGD01000141 JAC88748.1 EIM52653.1 KRT00337.1 ADMH02000712 ETN65262.1 EIM52654.2 OUUW01000007 SPP82810.1 SPP82808.1 SPP82809.1 GDHC01016465 JAQ02164.1 GDHC01018872 GDHC01010970 JAP99756.1 JAQ07659.1 DS235777 EEB16856.1 APGK01017248 KB739998 KB631984 ENN81985.1 ERL87674.1 GGFK01002789 MBW36110.1 GL732628 EFX69901.1 GDIQ01143712 JAL08014.1 GDIQ01080230 JAN14507.1 GDIP01166720 JAJ56682.1 AAAB01008847 EAA06889.5 GDIQ01017726 JAN77011.1 GDIP01135225 LRGB01003257 JAL68489.1 KZS03498.1 AXCM01001167 GFXV01000884 MBW12689.1 GDIP01165215 JAJ58187.1 ABLF02022457 GGMS01010893 MBY80096.1 GDIQ01034397 JAN60340.1 GDIQ01052105 JAN42632.1 GEFM01003084 JAP72712.1 GANP01006876 JAB77592.1 GDIQ01069884 JAN24853.1 GGFM01002050 MBW22801.1 GALX01005080 JAB63386.1 GGFK01002705 MBW36026.1 GDHF01009434 JAI42880.1 GDHF01023323 JAI28991.1 GDHF01014061 JAI38253.1 GFAA01003116 JAU00319.1 KJ405465 AHX26697.1 PZQS01000006 PVD28317.1 GEDC01010185 JAS27113.1 AMQN01009893 KB306507 ELT99775.1 GEDC01022494 JAS14804.1 QCYY01001653 ROT76507.1 NIVC01001575 PAA66247.1 NIVC01001611 PAA65857.1 GDIQ01126969 JAL24757.1 GFDL01000260 JAV34785.1 GDHF01027965 GDHF01010354 JAI24349.1 JAI41960.1 GDHF01002583 JAI49731.1 GDHF01032385 GDHF01031824 GDHF01002730 GDHF01000018 JAI19929.1 JAI20490.1 JAI49584.1 JAI52296.1 GDHF01014884 JAI37430.1 KQ765605 OAD53874.1 KQ435878 KOX70039.1 KQ978023 KYM98097.1 GGFM01002173 MBW22924.1 KQ971312 EEZ98246.1 ADTU01016063 KQ976725 KYM76594.1 GBYB01001600 JAG71367.1 GBYB01001599 JAG71366.1 GECZ01002257 JAS67512.1 GECZ01022585 JAS47184.1 GEBQ01009766 JAT30211.1 GEBQ01016307 JAT23670.1 NEVH01021956 PNF18977.1 PNF18985.1 PNF18979.1 GEZM01034847 JAV83616.1 GDIP01203960 JAJ19442.1 APCN01000987 KK852827 KDR15406.1
KQ560828 KXJ80718.1 GFDL01002114 JAV32931.1 KQ434896 KZC10725.1 CH477296 EAT44388.1 GANO01001616 JAB58255.1 GFDL01002115 JAV32930.1 LBMM01002703 KMQ94458.1 KZ288466 PBC25438.1 KQ978957 KYN27267.1 KQ982813 KYQ50276.1 GL453466 EFN75980.1 GAKP01019656 GAKP01019654 JAC39296.1 CVRI01000069 CRL06988.1 GAKP01019655 GAKP01019653 JAC39299.1 ATLV01013545 KE524867 KFB38085.1 CH479179 EDW24504.1 CM000070 EDY68029.3 KK107109 EZA59148.1 GGFJ01000775 MBW49916.1 GFTR01008718 JAW07708.1 GBGD01000141 JAC88748.1 EIM52653.1 KRT00337.1 ADMH02000712 ETN65262.1 EIM52654.2 OUUW01000007 SPP82810.1 SPP82808.1 SPP82809.1 GDHC01016465 JAQ02164.1 GDHC01018872 GDHC01010970 JAP99756.1 JAQ07659.1 DS235777 EEB16856.1 APGK01017248 KB739998 KB631984 ENN81985.1 ERL87674.1 GGFK01002789 MBW36110.1 GL732628 EFX69901.1 GDIQ01143712 JAL08014.1 GDIQ01080230 JAN14507.1 GDIP01166720 JAJ56682.1 AAAB01008847 EAA06889.5 GDIQ01017726 JAN77011.1 GDIP01135225 LRGB01003257 JAL68489.1 KZS03498.1 AXCM01001167 GFXV01000884 MBW12689.1 GDIP01165215 JAJ58187.1 ABLF02022457 GGMS01010893 MBY80096.1 GDIQ01034397 JAN60340.1 GDIQ01052105 JAN42632.1 GEFM01003084 JAP72712.1 GANP01006876 JAB77592.1 GDIQ01069884 JAN24853.1 GGFM01002050 MBW22801.1 GALX01005080 JAB63386.1 GGFK01002705 MBW36026.1 GDHF01009434 JAI42880.1 GDHF01023323 JAI28991.1 GDHF01014061 JAI38253.1 GFAA01003116 JAU00319.1 KJ405465 AHX26697.1 PZQS01000006 PVD28317.1 GEDC01010185 JAS27113.1 AMQN01009893 KB306507 ELT99775.1 GEDC01022494 JAS14804.1 QCYY01001653 ROT76507.1 NIVC01001575 PAA66247.1 NIVC01001611 PAA65857.1 GDIQ01126969 JAL24757.1 GFDL01000260 JAV34785.1 GDHF01027965 GDHF01010354 JAI24349.1 JAI41960.1 GDHF01002583 JAI49731.1 GDHF01032385 GDHF01031824 GDHF01002730 GDHF01000018 JAI19929.1 JAI20490.1 JAI49584.1 JAI52296.1 GDHF01014884 JAI37430.1 KQ765605 OAD53874.1 KQ435878 KOX70039.1 KQ978023 KYM98097.1 GGFM01002173 MBW22924.1 KQ971312 EEZ98246.1 ADTU01016063 KQ976725 KYM76594.1 GBYB01001600 JAG71367.1 GBYB01001599 JAG71366.1 GECZ01002257 JAS67512.1 GECZ01022585 JAS47184.1 GEBQ01009766 JAT30211.1 GEBQ01016307 JAT23670.1 NEVH01021956 PNF18977.1 PNF18985.1 PNF18979.1 GEZM01034847 JAV83616.1 GDIP01203960 JAJ19442.1 APCN01000987 KK852827 KDR15406.1
Proteomes
UP000007151
UP000053268
UP000069940
UP000249989
UP000076502
UP000008820
+ More
UP000036403 UP000242457 UP000078492 UP000075809 UP000008237 UP000091820 UP000183832 UP000069272 UP000030765 UP000008744 UP000001819 UP000053097 UP000000673 UP000268350 UP000009046 UP000019118 UP000030742 UP000000305 UP000007062 UP000076858 UP000075883 UP000007819 UP000245119 UP000014760 UP000076420 UP000283509 UP000215902 UP000005203 UP000053105 UP000078542 UP000007266 UP000005205 UP000078540 UP000235965 UP000075903 UP000075840 UP000027135
UP000036403 UP000242457 UP000078492 UP000075809 UP000008237 UP000091820 UP000183832 UP000069272 UP000030765 UP000008744 UP000001819 UP000053097 UP000000673 UP000268350 UP000009046 UP000019118 UP000030742 UP000000305 UP000007062 UP000076858 UP000075883 UP000007819 UP000245119 UP000014760 UP000076420 UP000283509 UP000215902 UP000005203 UP000053105 UP000078542 UP000007266 UP000005205 UP000078540 UP000235965 UP000075903 UP000075840 UP000027135
Pfam
Interpro
IPR000048
IQ_motif_EF-hand-BS
+ More
IPR036114 MYSc_Myo6
IPR032412 Myosin-VI_CBD
IPR027417 P-loop_NTPase
IPR036961 Kinesin_motor_dom_sf
IPR001609 Myosin_head_motor_dom
IPR019358 NEMP_fam
IPR020846 MFS_dom
IPR004009 Myosin_N
IPR013087 Znf_C2H2_type
IPR000210 BTB/POZ_dom
IPR011333 SKP1/BTB/POZ_sf
IPR008989 Myosin_S1_N
IPR036114 MYSc_Myo6
IPR032412 Myosin-VI_CBD
IPR027417 P-loop_NTPase
IPR036961 Kinesin_motor_dom_sf
IPR001609 Myosin_head_motor_dom
IPR019358 NEMP_fam
IPR020846 MFS_dom
IPR004009 Myosin_N
IPR013087 Znf_C2H2_type
IPR000210 BTB/POZ_dom
IPR011333 SKP1/BTB/POZ_sf
IPR008989 Myosin_S1_N
Gene 3D
ProteinModelPortal
A0A212EJV6
A0A194QD44
A0A1S4F6Z8
A0A182H7Y9
A0A1Q3FZD7
A0A154PFV1
+ More
A0A1S4F759 Q17DE9 U5EW23 A0A1Q3FZH6 A0A0J7KVX9 A0A2A3E163 A0A195EHC0 A0A151WQY6 E2C7U5 A0A1A9W6M3 A0A034V7N4 A0A1J1J5A6 A0A182FK19 A0A034V9W9 A0A084VJE1 B4G4F6 B5DYV4 A0A026WT79 A0A2M4BA63 A0A224XD56 A0A069DXG8 I5ANW8 W5JLL4 I5ANW9 A0A3B0K493 A0A3B0KGM1 A0A3B0K9Q5 A0A146L0Q7 A0A146LJJ8 E0VU00 N6UJR9 A0A2M4A5V3 E9HEA9 A0A0P5NPA2 A0A0P6DBF0 A0A0P5D452 Q7QE85 A0A0P6HLF0 A0A0P5SU58 A0A182MI83 A0A2H8TF30 A0A0P5D5E0 J9JYL7 A0A2S2QQP9 A0A0P6GI71 A0A0P6F7L5 A0A131Y2K9 V5HPH1 A0A0N8E1X3 A0A2M3Z2Q6 V5I894 A0A2M4A5D9 A0A0K8VWF7 A0A0K8UQX8 A0A0K8VHC1 A0A1E1XM31 A0A023PNK0 A0A2T7P4G3 A0A1B6DN64 R7U9G6 A0A1B6CMV6 A0A2C9JLB9 A0A3R7N3Q3 A0A267EZ74 A0A267EY54 A0A0P5Q1N5 A0A1Q3G4Q3 A0A088AJQ9 A0A0K8VTR8 A0A0K8WF13 A0A0K8WMD2 A0A0K8VFU5 A0A310S7H3 A0A0M8ZUX6 A0A195CBB8 A0A2M3Z322 D6W8W8 A0A158NHU4 A0A195AXB1 A0A158NHU5 A0A0C9R0T8 A0A0C9QE67 A0A1B6GYM3 A0A1B6FAM9 A0A1B6M2P4 A0A1B6LJ37 A0A2J7PRL7 A0A2J7PRL6 A0A2J7PRN3 A0A1Y1MI87 A0A0P5A5E9 A0A182VBR0 A0A182I9K4 A0A067QY55
A0A1S4F759 Q17DE9 U5EW23 A0A1Q3FZH6 A0A0J7KVX9 A0A2A3E163 A0A195EHC0 A0A151WQY6 E2C7U5 A0A1A9W6M3 A0A034V7N4 A0A1J1J5A6 A0A182FK19 A0A034V9W9 A0A084VJE1 B4G4F6 B5DYV4 A0A026WT79 A0A2M4BA63 A0A224XD56 A0A069DXG8 I5ANW8 W5JLL4 I5ANW9 A0A3B0K493 A0A3B0KGM1 A0A3B0K9Q5 A0A146L0Q7 A0A146LJJ8 E0VU00 N6UJR9 A0A2M4A5V3 E9HEA9 A0A0P5NPA2 A0A0P6DBF0 A0A0P5D452 Q7QE85 A0A0P6HLF0 A0A0P5SU58 A0A182MI83 A0A2H8TF30 A0A0P5D5E0 J9JYL7 A0A2S2QQP9 A0A0P6GI71 A0A0P6F7L5 A0A131Y2K9 V5HPH1 A0A0N8E1X3 A0A2M3Z2Q6 V5I894 A0A2M4A5D9 A0A0K8VWF7 A0A0K8UQX8 A0A0K8VHC1 A0A1E1XM31 A0A023PNK0 A0A2T7P4G3 A0A1B6DN64 R7U9G6 A0A1B6CMV6 A0A2C9JLB9 A0A3R7N3Q3 A0A267EZ74 A0A267EY54 A0A0P5Q1N5 A0A1Q3G4Q3 A0A088AJQ9 A0A0K8VTR8 A0A0K8WF13 A0A0K8WMD2 A0A0K8VFU5 A0A310S7H3 A0A0M8ZUX6 A0A195CBB8 A0A2M3Z322 D6W8W8 A0A158NHU4 A0A195AXB1 A0A158NHU5 A0A0C9R0T8 A0A0C9QE67 A0A1B6GYM3 A0A1B6FAM9 A0A1B6M2P4 A0A1B6LJ37 A0A2J7PRL7 A0A2J7PRL6 A0A2J7PRN3 A0A1Y1MI87 A0A0P5A5E9 A0A182VBR0 A0A182I9K4 A0A067QY55
PDB
2BKI
E-value=0,
Score=2283
Ontologies
GO
Topology
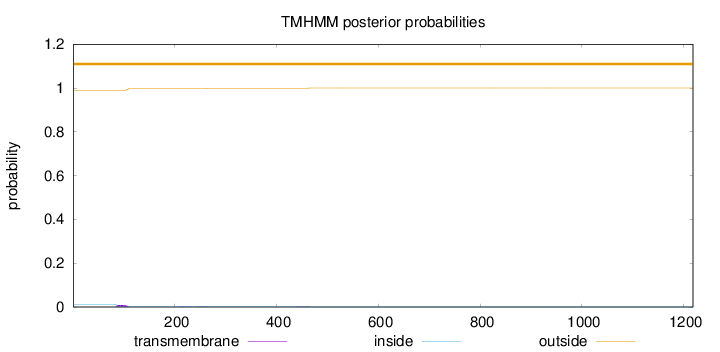
Length:
1218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23706
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01118
outside
1 - 1218
Population Genetic Test Statistics
Pi
245.657555
Theta
185.159301
Tajima's D
0.725138
CLR
0.722413
CSRT
0.578571071446428
Interpretation
Uncertain
