Gene
KWMTBOMO04875 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008050
Annotation
transgelin_[Antheraea_yamamai]
Full name
Transgelin
+ More
Calponin
Calponin
Location in the cell
Cytoplasmic Reliability : 2.036 Nuclear Reliability : 1.934
Sequence
CDS
ATGGCCGAGTATCGCGCAGGAAAAGCTGGCATCAACGCCGAAGCCCAGGCCAGGATCCACAGTAAATACAACGATGAAATCGCCCAAGAAACCCTCGAGTGGATCAGGAAATTGACCGGCGAACCGGCCAACACCTCCGGTGACGCCGACAACCTGTACGAAGTCTTAAAAGACGGCACCCTCCTCTGCAAACTAGTCAACGAGCTTCAACCAGGTTCCGTTAAGAAGATCAACCAAACGACCATGGCCTTCAAGTGTATGGAGAACATAAACGCTTTCCTAGAGGCAGTCAAGAAGCTAGGCGTGCCCGCTCAGGAGACCTTCCAGACCATTGATCTTTGGGAGAGGCAGAACCTTTACTCGGTGGTCGTTTGTCTACAGTCCCTCGGTAGGAAGGCAGGTAACTTCGGCAAGCCGTCCATCGGGCCCAAGGAGGCGGACAAGAATGTGCGCGACTTCTCCGAGGAGCAGCTGAAAGCCGGTCAGAACGTGATATCGCTACAGTACGGCACTAACAAGGGCCAGCAGTCGGGCATATCCTTCGGCAACAGGCGTCAAATGTGA
Protein
MAEYRAGKAGINAEAQARIHSKYNDEIAQETLEWIRKLTGEPANTSGDADNLYEVLKDGTLLCKLVNELQPGSVKKINQTTMAFKCMENINAFLEAVKKLGVPAQETFQTIDLWERQNLYSVVVCLQSLGRKAGNFGKPSIGPKEADKNVRDFSEEQLKAGQNVISLQYGTNKGQQSGISFGNRRQM
Summary
Description
Thin filament-associated protein that is implicated in the regulation and modulation of smooth muscle contraction. It is capable of binding to actin, calmodulin, troponin C and tropomyosin. The interaction of calponin with actin inhibits the actomyosin Mg-ATPase activity.
Similarity
Belongs to the calponin family.
Uniprot
H9JEV3
G8FVP3
A0A2A4ISJ9
A0A2H1X2F6
A0A194QCW4
A0A0L7KN16
+ More
A0A194QM03 A0A2W1BSE2 A0A1E1WIV4 A0A0K8SXC1 F8R4Z6 A0A212EJR1 A0A3S2TPH9 A0A1E1WN47 A0A194QTM3 I4DNE6 A0A212FDZ4 S4PMM4 Q1HQ70 A0A2A4J0G7 A0A2H1VJF7 B4L9E1 A0A2A3EBY9 V9IDZ7 A0A088AJ47 B3M4V0 A0A1W4W6W8 W8BCE9 A0A195EZC6 Q29D85 B4H7U8 A0A1B0GIN7 A0A3B0J6R8 B4LB83 A0A0M3QWF1 B4N3J5 A0A3L8DZH0 A0A158NDR9 A0A0C9QLT0 A0A0K8TZF1 A0A034WQN6 A0A1B6FMU2 A0A1L8EIE7 A0A0L0BR69 A0A1B6IPQ6 B4J1D9 A0A0J9RP62 B4PHP6 B4HU00 B3NG36 Q9VZI1 D3TSC5 A0A1B6L9J9 A0A0A1XN87 A0A1W4W8P8 A0A1I8P873 A0A1I8P874 R4UW18 A0A1L8E5Z6 A0A1I8NH87 U5EYQ9 A0A0A9WIH3 R4WR80 A0A023F9W3 A0A182NQJ8 V5GZ80 A0A170ZFV1 K7ISL6 A0A182RNK3 A0A0Q9XGY5 A0A1B6CS28 A0A182Q8Q7 A0A182VVS7 A0A182X9L4 A0A182VDH7 A0A182HKJ7 Q7PXA9 A0A069DPP4 T1DIU2 A0A336KB30 H9JC47 A0A0N8P109 A0A1J1II95 T1DN20 A0A2M3Z084 A0A2M4A8L1 A0A087ZJX0 A0A1L8EEX8 A0A1B0GQB3 A0A0L7R7U5 A0A1Q3EUJ0 A0A0R3P732 A0A0Q9WT59 A0A0K8TPC4 A0A224XXM0 A0A3B0JVN5 A0A0Q9WSE1 W8BSV8 Q1HR19 A0A023EJ28
A0A194QM03 A0A2W1BSE2 A0A1E1WIV4 A0A0K8SXC1 F8R4Z6 A0A212EJR1 A0A3S2TPH9 A0A1E1WN47 A0A194QTM3 I4DNE6 A0A212FDZ4 S4PMM4 Q1HQ70 A0A2A4J0G7 A0A2H1VJF7 B4L9E1 A0A2A3EBY9 V9IDZ7 A0A088AJ47 B3M4V0 A0A1W4W6W8 W8BCE9 A0A195EZC6 Q29D85 B4H7U8 A0A1B0GIN7 A0A3B0J6R8 B4LB83 A0A0M3QWF1 B4N3J5 A0A3L8DZH0 A0A158NDR9 A0A0C9QLT0 A0A0K8TZF1 A0A034WQN6 A0A1B6FMU2 A0A1L8EIE7 A0A0L0BR69 A0A1B6IPQ6 B4J1D9 A0A0J9RP62 B4PHP6 B4HU00 B3NG36 Q9VZI1 D3TSC5 A0A1B6L9J9 A0A0A1XN87 A0A1W4W8P8 A0A1I8P873 A0A1I8P874 R4UW18 A0A1L8E5Z6 A0A1I8NH87 U5EYQ9 A0A0A9WIH3 R4WR80 A0A023F9W3 A0A182NQJ8 V5GZ80 A0A170ZFV1 K7ISL6 A0A182RNK3 A0A0Q9XGY5 A0A1B6CS28 A0A182Q8Q7 A0A182VVS7 A0A182X9L4 A0A182VDH7 A0A182HKJ7 Q7PXA9 A0A069DPP4 T1DIU2 A0A336KB30 H9JC47 A0A0N8P109 A0A1J1II95 T1DN20 A0A2M3Z084 A0A2M4A8L1 A0A087ZJX0 A0A1L8EEX8 A0A1B0GQB3 A0A0L7R7U5 A0A1Q3EUJ0 A0A0R3P732 A0A0Q9WT59 A0A0K8TPC4 A0A224XXM0 A0A3B0JVN5 A0A0Q9WSE1 W8BSV8 Q1HR19 A0A023EJ28
Pubmed
19121390
26354079
26227816
28756777
21625546
22118469
+ More
22651552 23622113 17994087 24495485 15632085 30249741 21347285 25348373 26108605 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 25830018 25315136 25401762 26823975 23691247 25474469 20075255 12364791 14747013 17210077 26334808 24330624 20920257 26369729 17204158 17510324 24945155
22651552 23622113 17994087 24495485 15632085 30249741 21347285 25348373 26108605 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 25830018 25315136 25401762 26823975 23691247 25474469 20075255 12364791 14747013 17210077 26334808 24330624 20920257 26369729 17204158 17510324 24945155
EMBL
BABH01023365
JN082713
AER27811.1
NWSH01007453
PCG62967.1
ODYU01012876
+ More
SOQ59396.1 KQ459185 KPJ03383.1 JTDY01008557 KOB64515.1 KQ461198 KPJ05985.1 KZ149969 PZC76107.1 GDQN01004263 JAT86791.1 GBRD01007873 JAG57948.1 HM490090 ADM47613.1 AGBW02014406 OWR41727.1 RSAL01000031 RVE51633.1 GDQN01002778 JAT88276.1 KQ461137 KPJ08817.1 AK402870 KQ459603 BAM19436.1 KPI93065.1 AGBW02008995 OWR51940.1 GAIX01000101 JAA92459.1 DQ443182 ABF51271.1 NWSH01003936 PCG65657.1 ODYU01002898 SOQ40965.1 CH933816 EDW17316.1 KZ288290 PBC29238.1 JR041079 AEY59353.1 CH902618 EDV40524.1 GAMC01010188 JAB96367.1 KQ981906 KYN33262.1 CH379070 EAL30529.2 CH479219 EDW34738.1 AJWK01015120 AJWK01015121 AJWK01015122 OUUW01000002 SPP77425.1 CH940647 EDW68647.1 CP012525 ALC44039.1 CH964095 EDW79200.1 QOIP01000002 RLU25329.1 ADTU01012849 ADTU01012850 ADTU01012851 GBYB01004509 GBYB01011628 JAG74276.1 JAG81395.1 GDHF01032661 JAI19653.1 GAKP01002447 JAC56505.1 GECZ01018242 JAS51527.1 GFDG01000374 JAV18425.1 JRES01001494 KNC22483.1 GECU01018823 JAS88883.1 CH916366 EDV95830.1 CM002912 KMY97497.1 CM000159 EDW93355.1 CH480817 EDW50421.1 CH954178 EDV50865.1 AF217286 AE014296 AY060355 AAF44064.1 AAF47840.2 AAL25394.1 EZ424327 ADD20603.1 GEBQ01019588 JAT20389.1 GBXI01002244 JAD12048.1 KC571917 AGM32416.1 GFDF01000152 JAV13932.1 GANO01001867 JAB58004.1 GBHO01036398 GBRD01016895 GDHC01000434 JAG07206.1 JAG48932.1 JAQ18195.1 AK417172 BAN20387.1 GBBI01000923 JAC17789.1 GALX01001249 JAB67217.1 GEMB01002249 JAS00931.1 KRG07727.1 GEDC01021255 JAS16043.1 AXCN02001518 APCN01000834 AAAB01008987 EAA01312.4 GBGD01003099 JAC85790.1 GALA01000756 JAA94096.1 UFQS01000192 UFQT01000192 SSX01032.1 SSX21412.1 BABH01014894 KPU78612.1 CVRI01000048 CRK98774.1 GAMD01003245 JAA98345.1 GGFM01001184 MBW21935.1 GGFK01003815 MBW37136.1 GGFL01004380 MBW68558.1 GFDG01001551 JAV17248.1 AJVK01036080 KQ414637 KOC66950.1 GFDL01016119 JAV18926.1 KRT09031.1 KRF83950.1 GDAI01001366 JAI16237.1 GFTR01003134 JAW13292.1 SPP77426.1 KRF99024.1 GAMC01010189 JAB96366.1 DQ440275 CH477412 ABF18308.1 EAT41438.1 GAPW01004775 GAPW01004774 JAC08824.1
SOQ59396.1 KQ459185 KPJ03383.1 JTDY01008557 KOB64515.1 KQ461198 KPJ05985.1 KZ149969 PZC76107.1 GDQN01004263 JAT86791.1 GBRD01007873 JAG57948.1 HM490090 ADM47613.1 AGBW02014406 OWR41727.1 RSAL01000031 RVE51633.1 GDQN01002778 JAT88276.1 KQ461137 KPJ08817.1 AK402870 KQ459603 BAM19436.1 KPI93065.1 AGBW02008995 OWR51940.1 GAIX01000101 JAA92459.1 DQ443182 ABF51271.1 NWSH01003936 PCG65657.1 ODYU01002898 SOQ40965.1 CH933816 EDW17316.1 KZ288290 PBC29238.1 JR041079 AEY59353.1 CH902618 EDV40524.1 GAMC01010188 JAB96367.1 KQ981906 KYN33262.1 CH379070 EAL30529.2 CH479219 EDW34738.1 AJWK01015120 AJWK01015121 AJWK01015122 OUUW01000002 SPP77425.1 CH940647 EDW68647.1 CP012525 ALC44039.1 CH964095 EDW79200.1 QOIP01000002 RLU25329.1 ADTU01012849 ADTU01012850 ADTU01012851 GBYB01004509 GBYB01011628 JAG74276.1 JAG81395.1 GDHF01032661 JAI19653.1 GAKP01002447 JAC56505.1 GECZ01018242 JAS51527.1 GFDG01000374 JAV18425.1 JRES01001494 KNC22483.1 GECU01018823 JAS88883.1 CH916366 EDV95830.1 CM002912 KMY97497.1 CM000159 EDW93355.1 CH480817 EDW50421.1 CH954178 EDV50865.1 AF217286 AE014296 AY060355 AAF44064.1 AAF47840.2 AAL25394.1 EZ424327 ADD20603.1 GEBQ01019588 JAT20389.1 GBXI01002244 JAD12048.1 KC571917 AGM32416.1 GFDF01000152 JAV13932.1 GANO01001867 JAB58004.1 GBHO01036398 GBRD01016895 GDHC01000434 JAG07206.1 JAG48932.1 JAQ18195.1 AK417172 BAN20387.1 GBBI01000923 JAC17789.1 GALX01001249 JAB67217.1 GEMB01002249 JAS00931.1 KRG07727.1 GEDC01021255 JAS16043.1 AXCN02001518 APCN01000834 AAAB01008987 EAA01312.4 GBGD01003099 JAC85790.1 GALA01000756 JAA94096.1 UFQS01000192 UFQT01000192 SSX01032.1 SSX21412.1 BABH01014894 KPU78612.1 CVRI01000048 CRK98774.1 GAMD01003245 JAA98345.1 GGFM01001184 MBW21935.1 GGFK01003815 MBW37136.1 GGFL01004380 MBW68558.1 GFDG01001551 JAV17248.1 AJVK01036080 KQ414637 KOC66950.1 GFDL01016119 JAV18926.1 KRT09031.1 KRF83950.1 GDAI01001366 JAI16237.1 GFTR01003134 JAW13292.1 SPP77426.1 KRF99024.1 GAMC01010189 JAB96366.1 DQ440275 CH477412 ABF18308.1 EAT41438.1 GAPW01004775 GAPW01004774 JAC08824.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000037510
UP000053240
UP000007151
+ More
UP000283053 UP000009192 UP000242457 UP000005203 UP000007801 UP000192221 UP000078541 UP000001819 UP000008744 UP000092461 UP000268350 UP000008792 UP000092553 UP000007798 UP000279307 UP000005205 UP000037069 UP000001070 UP000002282 UP000001292 UP000008711 UP000000803 UP000192223 UP000095300 UP000095301 UP000075884 UP000002358 UP000075900 UP000075886 UP000075920 UP000076407 UP000075903 UP000075840 UP000007062 UP000183832 UP000092462 UP000053825 UP000008820
UP000283053 UP000009192 UP000242457 UP000005203 UP000007801 UP000192221 UP000078541 UP000001819 UP000008744 UP000092461 UP000268350 UP000008792 UP000092553 UP000007798 UP000279307 UP000005205 UP000037069 UP000001070 UP000002282 UP000001292 UP000008711 UP000000803 UP000192223 UP000095300 UP000095301 UP000075884 UP000002358 UP000075900 UP000075886 UP000075920 UP000076407 UP000075903 UP000075840 UP000007062 UP000183832 UP000092462 UP000053825 UP000008820
Interpro
SUPFAM
SSF47576
SSF47576
Gene 3D
CDD
ProteinModelPortal
H9JEV3
G8FVP3
A0A2A4ISJ9
A0A2H1X2F6
A0A194QCW4
A0A0L7KN16
+ More
A0A194QM03 A0A2W1BSE2 A0A1E1WIV4 A0A0K8SXC1 F8R4Z6 A0A212EJR1 A0A3S2TPH9 A0A1E1WN47 A0A194QTM3 I4DNE6 A0A212FDZ4 S4PMM4 Q1HQ70 A0A2A4J0G7 A0A2H1VJF7 B4L9E1 A0A2A3EBY9 V9IDZ7 A0A088AJ47 B3M4V0 A0A1W4W6W8 W8BCE9 A0A195EZC6 Q29D85 B4H7U8 A0A1B0GIN7 A0A3B0J6R8 B4LB83 A0A0M3QWF1 B4N3J5 A0A3L8DZH0 A0A158NDR9 A0A0C9QLT0 A0A0K8TZF1 A0A034WQN6 A0A1B6FMU2 A0A1L8EIE7 A0A0L0BR69 A0A1B6IPQ6 B4J1D9 A0A0J9RP62 B4PHP6 B4HU00 B3NG36 Q9VZI1 D3TSC5 A0A1B6L9J9 A0A0A1XN87 A0A1W4W8P8 A0A1I8P873 A0A1I8P874 R4UW18 A0A1L8E5Z6 A0A1I8NH87 U5EYQ9 A0A0A9WIH3 R4WR80 A0A023F9W3 A0A182NQJ8 V5GZ80 A0A170ZFV1 K7ISL6 A0A182RNK3 A0A0Q9XGY5 A0A1B6CS28 A0A182Q8Q7 A0A182VVS7 A0A182X9L4 A0A182VDH7 A0A182HKJ7 Q7PXA9 A0A069DPP4 T1DIU2 A0A336KB30 H9JC47 A0A0N8P109 A0A1J1II95 T1DN20 A0A2M3Z084 A0A2M4A8L1 A0A087ZJX0 A0A1L8EEX8 A0A1B0GQB3 A0A0L7R7U5 A0A1Q3EUJ0 A0A0R3P732 A0A0Q9WT59 A0A0K8TPC4 A0A224XXM0 A0A3B0JVN5 A0A0Q9WSE1 W8BSV8 Q1HR19 A0A023EJ28
A0A194QM03 A0A2W1BSE2 A0A1E1WIV4 A0A0K8SXC1 F8R4Z6 A0A212EJR1 A0A3S2TPH9 A0A1E1WN47 A0A194QTM3 I4DNE6 A0A212FDZ4 S4PMM4 Q1HQ70 A0A2A4J0G7 A0A2H1VJF7 B4L9E1 A0A2A3EBY9 V9IDZ7 A0A088AJ47 B3M4V0 A0A1W4W6W8 W8BCE9 A0A195EZC6 Q29D85 B4H7U8 A0A1B0GIN7 A0A3B0J6R8 B4LB83 A0A0M3QWF1 B4N3J5 A0A3L8DZH0 A0A158NDR9 A0A0C9QLT0 A0A0K8TZF1 A0A034WQN6 A0A1B6FMU2 A0A1L8EIE7 A0A0L0BR69 A0A1B6IPQ6 B4J1D9 A0A0J9RP62 B4PHP6 B4HU00 B3NG36 Q9VZI1 D3TSC5 A0A1B6L9J9 A0A0A1XN87 A0A1W4W8P8 A0A1I8P873 A0A1I8P874 R4UW18 A0A1L8E5Z6 A0A1I8NH87 U5EYQ9 A0A0A9WIH3 R4WR80 A0A023F9W3 A0A182NQJ8 V5GZ80 A0A170ZFV1 K7ISL6 A0A182RNK3 A0A0Q9XGY5 A0A1B6CS28 A0A182Q8Q7 A0A182VVS7 A0A182X9L4 A0A182VDH7 A0A182HKJ7 Q7PXA9 A0A069DPP4 T1DIU2 A0A336KB30 H9JC47 A0A0N8P109 A0A1J1II95 T1DN20 A0A2M3Z084 A0A2M4A8L1 A0A087ZJX0 A0A1L8EEX8 A0A1B0GQB3 A0A0L7R7U5 A0A1Q3EUJ0 A0A0R3P732 A0A0Q9WT59 A0A0K8TPC4 A0A224XXM0 A0A3B0JVN5 A0A0Q9WSE1 W8BSV8 Q1HR19 A0A023EJ28
PDB
1WYM
E-value=4.27899e-17,
Score=211
Ontologies
GO
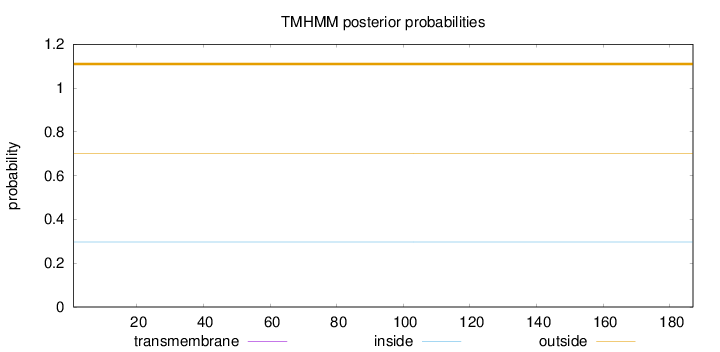
Topology
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.29737
outside
1 - 187
Population Genetic Test Statistics
Pi
211.056842
Theta
208.080043
Tajima's D
0.045037
CLR
0
CSRT
0.384380780960952
Interpretation
Uncertain
