Gene
KWMTBOMO04873 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008023
Annotation
hypothetical_protein_RR46_06541_[Papilio_xuthus]
Full name
Kynurenine 3-monooxygenase
Alternative Name
Kynurenine 3-hydroxylase
Location in the cell
Cytoplasmic Reliability : 1.793 Nuclear Reliability : 1.393
Sequence
CDS
ATGTTATGTTATCCGACAGACGCAAGGAGCTGTGTGACGTGCGACAAGGTGCTGGATGAGTTGGAGAAGATCGACGACGACACGGACACGTTCGGCGTGGACTTCGTCAAGATCAACGACAAGCGGCTCGCGAAGCAGTATGGCATCACGAAATTCCCCGCCCTCACGTACTTCCGTGAGAAGGAACCGATCATTTACGAAGGAGATCTCATGGACGAAGAAAGTGTCCTGGATTTCCTGACGAGCTTAGAAGCAATGGACCTTCCTGACCGGATAGAAGAGGTTAACCAAAAGATTCTGTCGAAAATTATCGAGGAGACAGACTATGTAGCAGTGCTTTTCTGTCCGGATCACAAAAGTTGCGGTCCCTCAAACAACAAGCCGGAGTGTAAGAAATGTGCGAAGGCTCTACAAGAGTTGGAGAACATTGACGACGAGGCCGACCAGCTCGGGATTGGTTTCGTGAAGATCCACGACGAGGAATTGGCCGAGGAGTACAGTCTTGGAGAACTGCCAAGACTGGTCTACTATAGGCATCAAATACCTATTATCTATGAAGGTGAATTGAGCAAGGAAGAGGACGTGCTGGAGTGGCTGATTGCCAACAAGTCTACCGGCGATGAGGAGGACGTCATCGAAGACGTAACAGCCAAGACCTTGAATACTCTGATTATCAACGTCGATCATCTTGTTGTACTCTTTTACGACCACGGCGACGAAGAATCAATGACTGTTCTGAATGAGTTGGAGAAGATAGACGATGACTGCGACCGTCACGGGATCCAGTTCGTCAAGATCGACGACCAGAAGGCGGCGCGAGCTTTCGGCATTGATGACCTACCCTCCATAGTCTACTTCGAGAAGCAAATACCAAATGTTTATGACGGAGACCTTGAAAACGAAGAAGAAATCTTAGAGTGGCTTGTAGACCAACTAGAGAAGGATGAAATTGAAGATGTCACCGATGAAATGCTGGACCGGCTCATCAAGGACGGGAAAACTGTCGCCGTGTTGTTTTATGACAACAACGATCGTAGGTCACAGAAAGTGCTCAACGAATTGGAAAATATCGATGACGAATGCGATCAGCTCGGCATTGCGTTTGTGAAAATTGACAACGACGAGGAAGCCAAAGAATACGGCATCGAAAAGATACCGACGCTTTTGTACTTTGAAAAGGGAATACCCACGTACTACGAAGGAAACCTTGAAGAAGAGGAAAAAGTATTAGAATGGTTGCAACATCAAGCCGAGAGTGACGAAATCGAAGACATCACCGACGAGATGCTGGACCTCATCATTGAGAAGATGCCGTACGTGGCCGTCTTGTTCTACGACAAGGACCAAAAGAAAAGCCAAAAGGTCCTGGTTGAGTTGGAGAACATTGATGATGAATGCGATCAAAATGACATCGCCTTCGTGAAAATAGATGACGACAAAGAGGCCAAAGAGTATGGCATTGAAACCATCCCTACCATGGTGTTCTTTGAAAAGGGTATCCCACATATTTATGAAGGTGATTTGATGAAGGAAGAAGAGCTTCTTGGATGGTTGCTGCATCAGAAACGGCACAGTGAAATCCCTGAAGTCACGGACGAAATGATGGACAAACTCATCGAAACAACTCCTTATTTGGCGGTTATTTTCTATGACAAAGACGACAAGCAAGACATCAGGATCCTCAATGAACTTGAGAACATCGACGATGAATTAGAGAAAGAAGGCATCGTTATTGTGCGTATGGACAACGATGCAGAAGCCAAAGAATACGGCATAGATCATCTGCCCACGCTGGTCTACTTCGAGGAGCATATCCCGGCGATATACGAAGGCGACCTCATGAACGAAGACGAAGTGCTGGAATGGCTGATCGAACAGAAGAACAGCGCGACCATCGAGGAAGTCACCGATGAGATATTAGTCGATCTTATTGAAGAGCACGAATATGTTGTCGTGTATTTCAGCGGTAACTGCGAGGAAGGCGAGGAATGCGACAACATCTTGGATGAACTGGAGAATATCGACGACGAGCTTGATGAGACCGGCATTATCTTCGTGACCACCGAGGACACCAGCTTAGCCAAGAAGTACGGCATCAAGACGTTCCCGACCCTCGTGTTCTTCAGGAACAAGGATCCGTTAGTCTACAAAGGTGACATCGAAGACGAAGATGAAGTATTGGCGTGGTTAACTGACGAGAACACATTGGAAATTCCTGGCAAAATTGAAGAGGTCAATTCTAAAATGCTTGAAAAGATTCTAGAAGAGAACGATCACGTCGTTGTATTCTTCTACAAAGAAGGTGACAAGAAGTCTCAAAAGATTCTAAGCGAATTGGAAAACATTGACGACGAGTGTGAAGAAAAAGACATCGACTTTGTCAAGACATCAGACAACGGCATCGACAAGGAATACGACCTCCCAGAATTACCAGCCTTGGCTTTCTACAGGCACAAGTTCAGGACCATCTACGAAGGAGACTTGATGCACGAAGAAGCTATCTTAAAGTGGGTTTTAGATCTGCACGATTCACAACCGGATATCATTGAAAACGTCGACAGGAAGACTCTTAAGAACCTAATCAATGATGTTGAACATTTGGCCGTTTTCTTCTACAGTGAAGATTGTGACACGTGCGACGACATCCTGGTGGAGCTGGAGACTATTGATGACGACACTGACAAGCACGGCATCCAATTCGTAAAGTCAAAGGATTCTAAGCTAGCTACCGATATAGGAATATTCAGTTTCCCGGCTTTGGTTTATTACGAAACTGGTGTTCCGATTATGTACGACGGTGACTTAAAGAACGAAAATAAGGTTCTCCAGTGGCTTGTAGATCAAAAAAGCGATCGCTGTTTCTACATTGGATTGGGGGCGAAACCGGCCGTCCCAAAAATGACTTACGAACCATACCAGTGCTGTCCCACTAAAGTTCAGAGCCCGACTAAAGTAGCCAAGGCCACGCCGACCAAGGTGCCACCCAAGAAACTGGAGAAACAAGAAAAGGGAAAACCCAAACAGCAGGAAAAGGAAAAAGAAAAGCCGTCTAAAGGGAAAAATAAGGCACTCGCCAAGCCCGACAAACCTTCTAAAGGGAAAAAAGCTGAAGATAATTATGAAAACTTTTTAGGAGATCTTTTGGACGAATCTGAGGTACTCGACTGGATGGTGAAACAAAAAGAAGATGAAAGCATTGAAGAAATAGACAGAGATGAATTGTTCAAATATATCGAAACGAAAGAGTTTTTAGCCGTTATATTTTATAAAGAAGACGACCCAGAAAGCCCAAGAGTTCTGAGGCACGTGGAGCTGATAGACGACGAAGCGGCTCAGTACGGTATCAAAATCGCAAAATGCAGCGACAAATTGATGGCCAAGAAATACGGCTTCCGCAATCCGCCGGGCATTACCTATTTCAGAAAAACTAAATACATTAATTACGATGGCGACATTGACGACGAAGAAGAGATATTGGACTGGCTGACGAATCCAGAAAACATGGAGCTGACTGATCACATCGAGAAAGTTAATAGGAAAATGTTTCAGAAGATAAGACAAACGTCGGATTACGTTGCTGTGTTCTTTTGTGAGTGTGCAACATTTTTTATTTATTTTTTAATTGCGTAG
Protein
MLCYPTDARSCVTCDKVLDELEKIDDDTDTFGVDFVKINDKRLAKQYGITKFPALTYFREKEPIIYEGDLMDEESVLDFLTSLEAMDLPDRIEEVNQKILSKIIEETDYVAVLFCPDHKSCGPSNNKPECKKCAKALQELENIDDEADQLGIGFVKIHDEELAEEYSLGELPRLVYYRHQIPIIYEGELSKEEDVLEWLIANKSTGDEEDVIEDVTAKTLNTLIINVDHLVVLFYDHGDEESMTVLNELEKIDDDCDRHGIQFVKIDDQKAARAFGIDDLPSIVYFEKQIPNVYDGDLENEEEILEWLVDQLEKDEIEDVTDEMLDRLIKDGKTVAVLFYDNNDRRSQKVLNELENIDDECDQLGIAFVKIDNDEEAKEYGIEKIPTLLYFEKGIPTYYEGNLEEEEKVLEWLQHQAESDEIEDITDEMLDLIIEKMPYVAVLFYDKDQKKSQKVLVELENIDDECDQNDIAFVKIDDDKEAKEYGIETIPTMVFFEKGIPHIYEGDLMKEEELLGWLLHQKRHSEIPEVTDEMMDKLIETTPYLAVIFYDKDDKQDIRILNELENIDDELEKEGIVIVRMDNDAEAKEYGIDHLPTLVYFEEHIPAIYEGDLMNEDEVLEWLIEQKNSATIEEVTDEILVDLIEEHEYVVVYFSGNCEEGEECDNILDELENIDDELDETGIIFVTTEDTSLAKKYGIKTFPTLVFFRNKDPLVYKGDIEDEDEVLAWLTDENTLEIPGKIEEVNSKMLEKILEENDHVVVFFYKEGDKKSQKILSELENIDDECEEKDIDFVKTSDNGIDKEYDLPELPALAFYRHKFRTIYEGDLMHEEAILKWVLDLHDSQPDIIENVDRKTLKNLINDVEHLAVFFYSEDCDTCDDILVELETIDDDTDKHGIQFVKSKDSKLATDIGIFSFPALVYYETGVPIMYDGDLKNENKVLQWLVDQKSDRCFYIGLGAKPAVPKMTYEPYQCCPTKVQSPTKVAKATPTKVPPKKLEKQEKGKPKQQEKEKEKPSKGKNKALAKPDKPSKGKKAEDNYENFLGDLLDESEVLDWMVKQKEDESIEEIDRDELFKYIETKEFLAVIFYKEDDPESPRVLRHVELIDDEAAQYGIKIAKCSDKLMAKKYGFRNPPGITYFRKTKYINYDGDIDDEEEILDWLTNPENMELTDHIEKVNRKMFQKIRQTSDYVAVFFCECATFFIYFLIA
Summary
Description
Catalyzes the hydroxylation of L-kynurenine (L-Kyn) to form 3-hydroxy-L-kynurenine (L-3OHKyn). Required for synthesis of quinolinic acid.
Catalytic Activity
H(+) + L-kynurenine + NADPH + O2 = 3-hydroxy-L-kynurenine + H2O + NADP(+)
Cofactor
FAD
Similarity
Belongs to the aromatic-ring hydroxylase family. KMO subfamily.
Uniprot
A0A194QEL2
A0A0L7LBA3
A0A195FV70
A0A195B1Q4
E2BE86
F4WXX7
+ More
H9JES6 A0A067R7B8 A0A194QKC7 A0A2W1BR63 A0A2H1W476 A0A3S2LCA2 A0A0L7RGJ2 A0A212EJR9 A0A0C9RUC2 A0A3L8DY73 A0A026X3Q6 A0A151WTY0 A0A195CIJ3 A0A158NZ91 A0A0J9TUG9 K7IYV6 A0A1W4VFE8 A0A0Q9X0P2 A0A1B6EFX8 A0A1B6F054 A0A0J9TUG3 A0A0Q5WNY3 A0A1B6LBI4 A0A1B6DCF0 A0A0L0CE53 A0A232ERZ5 A0A1Y1L3H5 A0A1Y1KXP6 W8AVA6 W8AR81 B4P107 A0A1W4VE31 A0A1W4V1T0 B4HQB3 A0A0C9R9T6 B4J6K1 A0A139WMR8 A0A034VR99 A0A034VUQ8 A0A0M4E6M5 A0A139WM79 A0A1Y1L3G5 A0A0T6B0M2 E2AH21 A0A1Y1L4Y4 A0A2R7VRI3 A0A1Y1KXN3 A0A1Y1L045 A0A0J9TUH8 B3MIN9 Q9V9A9 A0A0J9R6U2 E1JGY6 A0A1I8P428 A0A1I8P422 A0A1I8P3Y7 A0A1W4VEI9 A0A1I8P433 B3NB27 A0A1W4VE25 B4LK14 A0A1W4V1S5 A0A1W4VFE3 B4MY49 A0A0Q9X1D6 A0A146M1J4 A0A0C9RFW1 A0A1I8M374 A0A1I8M384 A0A1I8M378 A0A1I8M379 A0A1W4WET8 A0A139WM70 A0A1Y1L171 A0A1W4W2X2 A0A034VRA7 A0A2J7PVL4 A0A1Y1L2F8 A0A034VUR2 A0A1A9UIW9 A0A1J1I7I3 A0A1J1IBB7 A0A0J9R6J4 A0A0J9R6I3 Q0E9N2 A0A1I8P3Y0 A0A0J9R6T3 A0A1I8P423 A0A1W4V279 U5EMW7 A0A1B0EXM3 A0A2H8TVP0 A0A1I8M376 A0A2J7PVK4
H9JES6 A0A067R7B8 A0A194QKC7 A0A2W1BR63 A0A2H1W476 A0A3S2LCA2 A0A0L7RGJ2 A0A212EJR9 A0A0C9RUC2 A0A3L8DY73 A0A026X3Q6 A0A151WTY0 A0A195CIJ3 A0A158NZ91 A0A0J9TUG9 K7IYV6 A0A1W4VFE8 A0A0Q9X0P2 A0A1B6EFX8 A0A1B6F054 A0A0J9TUG3 A0A0Q5WNY3 A0A1B6LBI4 A0A1B6DCF0 A0A0L0CE53 A0A232ERZ5 A0A1Y1L3H5 A0A1Y1KXP6 W8AVA6 W8AR81 B4P107 A0A1W4VE31 A0A1W4V1T0 B4HQB3 A0A0C9R9T6 B4J6K1 A0A139WMR8 A0A034VR99 A0A034VUQ8 A0A0M4E6M5 A0A139WM79 A0A1Y1L3G5 A0A0T6B0M2 E2AH21 A0A1Y1L4Y4 A0A2R7VRI3 A0A1Y1KXN3 A0A1Y1L045 A0A0J9TUH8 B3MIN9 Q9V9A9 A0A0J9R6U2 E1JGY6 A0A1I8P428 A0A1I8P422 A0A1I8P3Y7 A0A1W4VEI9 A0A1I8P433 B3NB27 A0A1W4VE25 B4LK14 A0A1W4V1S5 A0A1W4VFE3 B4MY49 A0A0Q9X1D6 A0A146M1J4 A0A0C9RFW1 A0A1I8M374 A0A1I8M384 A0A1I8M378 A0A1I8M379 A0A1W4WET8 A0A139WM70 A0A1Y1L171 A0A1W4W2X2 A0A034VRA7 A0A2J7PVL4 A0A1Y1L2F8 A0A034VUR2 A0A1A9UIW9 A0A1J1I7I3 A0A1J1IBB7 A0A0J9R6J4 A0A0J9R6I3 Q0E9N2 A0A1I8P3Y0 A0A0J9R6T3 A0A1I8P423 A0A1W4V279 U5EMW7 A0A1B0EXM3 A0A2H8TVP0 A0A1I8M376 A0A2J7PVK4
EC Number
1.14.13.9
Pubmed
26354079
26227816
20798317
21719571
19121390
24845553
+ More
28756777 22118469 30249741 24508170 21347285 22936249 20075255 17994087 26108605 28648823 28004739 24495485 17550304 18362917 19820115 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 26823975 25315136
28756777 22118469 30249741 24508170 21347285 22936249 20075255 17994087 26108605 28648823 28004739 24495485 17550304 18362917 19820115 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 26823975 25315136
EMBL
KQ459185
KPJ03385.1
JTDY01001873
KOB72664.1
KQ981276
KYN43764.1
+ More
KQ976681 KYM78212.1 GL447768 EFN85931.1 GL888434 EGI60913.1 BABH01023357 BABH01023358 BABH01023359 BABH01023360 BABH01023361 KK852827 KDR15395.1 KQ461198 KPJ05987.1 KZ149969 PZC76105.1 ODYU01006240 SOQ47899.1 RSAL01000048 RVE50479.1 KQ414596 KOC69939.1 AGBW02014406 OWR41726.1 GBYB01011201 JAG80968.1 QOIP01000003 RLU24899.1 KK107013 EZA62912.1 KQ982748 KYQ51296.1 KQ977696 KYN00550.1 ADTU01004077 ADTU01004078 CM002911 KMY91790.1 CH963894 KRF98566.1 GEDC01011253 GEDC01000460 JAS26045.1 JAS36838.1 GECZ01026147 JAS43622.1 KMY91785.1 CH954177 KQS71010.1 GEBQ01018942 JAT21035.1 GEDC01013935 GEDC01004680 JAS23363.1 JAS32618.1 JRES01000505 KNC30530.1 NNAY01002528 OXU21133.1 GEZM01071025 GEZM01071017 JAV66136.1 GEZM01071018 JAV66134.1 GAMC01017987 JAB88568.1 GAMC01017988 JAB88567.1 CM000157 EDW89081.2 CH480816 EDW46652.1 GBYB01009697 JAG79464.1 CH916367 EDW02002.1 KQ971312 KYB29137.1 GAKP01013136 GAKP01013128 JAC45816.1 GAKP01013125 GAKP01013121 JAC45827.1 CP012524 ALC42178.1 KYB29139.1 GEZM01071026 GEZM01071022 JAV66126.1 LJIG01016421 KRT80653.1 GL439460 EFN67268.1 GEZM01071021 GEZM01071016 JAV66137.1 KK854011 PTY09280.1 GEZM01071024 GEZM01071023 JAV66122.1 GEZM01071019 GEZM01071014 JAV66141.1 KMY91795.1 CH902619 EDV38115.2 AF216973 AE013599 AAF34746.1 AAN16126.1 KMY91791.1 ACZ94345.1 EDV59792.2 CH940648 EDW61668.2 EDW77038.2 KRF98564.1 KRF98565.1 KRF98567.1 GDHC01004946 JAQ13683.1 GBYB01007240 JAG77007.1 KYB29138.1 GEZM01071015 GEZM01071013 JAV66140.1 GAKP01013126 JAC45826.1 NEVH01020956 PNF20369.1 GEZM01071020 GEZM01071012 JAV66145.1 GAKP01013131 GAKP01013120 JAC45832.1 CVRI01000043 CRK96271.1 CRK96270.1 KMY91797.1 KMY91787.1 ABI31026.3 KMY91798.1 GANO01004322 JAB55549.1 AJWK01034186 GFXV01005413 MBW17218.1 PNF20363.1
KQ976681 KYM78212.1 GL447768 EFN85931.1 GL888434 EGI60913.1 BABH01023357 BABH01023358 BABH01023359 BABH01023360 BABH01023361 KK852827 KDR15395.1 KQ461198 KPJ05987.1 KZ149969 PZC76105.1 ODYU01006240 SOQ47899.1 RSAL01000048 RVE50479.1 KQ414596 KOC69939.1 AGBW02014406 OWR41726.1 GBYB01011201 JAG80968.1 QOIP01000003 RLU24899.1 KK107013 EZA62912.1 KQ982748 KYQ51296.1 KQ977696 KYN00550.1 ADTU01004077 ADTU01004078 CM002911 KMY91790.1 CH963894 KRF98566.1 GEDC01011253 GEDC01000460 JAS26045.1 JAS36838.1 GECZ01026147 JAS43622.1 KMY91785.1 CH954177 KQS71010.1 GEBQ01018942 JAT21035.1 GEDC01013935 GEDC01004680 JAS23363.1 JAS32618.1 JRES01000505 KNC30530.1 NNAY01002528 OXU21133.1 GEZM01071025 GEZM01071017 JAV66136.1 GEZM01071018 JAV66134.1 GAMC01017987 JAB88568.1 GAMC01017988 JAB88567.1 CM000157 EDW89081.2 CH480816 EDW46652.1 GBYB01009697 JAG79464.1 CH916367 EDW02002.1 KQ971312 KYB29137.1 GAKP01013136 GAKP01013128 JAC45816.1 GAKP01013125 GAKP01013121 JAC45827.1 CP012524 ALC42178.1 KYB29139.1 GEZM01071026 GEZM01071022 JAV66126.1 LJIG01016421 KRT80653.1 GL439460 EFN67268.1 GEZM01071021 GEZM01071016 JAV66137.1 KK854011 PTY09280.1 GEZM01071024 GEZM01071023 JAV66122.1 GEZM01071019 GEZM01071014 JAV66141.1 KMY91795.1 CH902619 EDV38115.2 AF216973 AE013599 AAF34746.1 AAN16126.1 KMY91791.1 ACZ94345.1 EDV59792.2 CH940648 EDW61668.2 EDW77038.2 KRF98564.1 KRF98565.1 KRF98567.1 GDHC01004946 JAQ13683.1 GBYB01007240 JAG77007.1 KYB29138.1 GEZM01071015 GEZM01071013 JAV66140.1 GAKP01013126 JAC45826.1 NEVH01020956 PNF20369.1 GEZM01071020 GEZM01071012 JAV66145.1 GAKP01013131 GAKP01013120 JAC45832.1 CVRI01000043 CRK96271.1 CRK96270.1 KMY91797.1 KMY91787.1 ABI31026.3 KMY91798.1 GANO01004322 JAB55549.1 AJWK01034186 GFXV01005413 MBW17218.1 PNF20363.1
Proteomes
UP000053268
UP000037510
UP000078541
UP000078540
UP000008237
UP000007755
+ More
UP000005204 UP000027135 UP000053240 UP000283053 UP000053825 UP000007151 UP000279307 UP000053097 UP000075809 UP000078542 UP000005205 UP000002358 UP000192221 UP000007798 UP000008711 UP000037069 UP000215335 UP000002282 UP000001292 UP000001070 UP000007266 UP000092553 UP000000311 UP000007801 UP000000803 UP000095300 UP000008792 UP000095301 UP000192223 UP000235965 UP000078200 UP000183832 UP000092461
UP000005204 UP000027135 UP000053240 UP000283053 UP000053825 UP000007151 UP000279307 UP000053097 UP000075809 UP000078542 UP000005205 UP000002358 UP000192221 UP000007798 UP000008711 UP000037069 UP000215335 UP000002282 UP000001292 UP000001070 UP000007266 UP000092553 UP000000311 UP000007801 UP000000803 UP000095300 UP000008792 UP000095301 UP000192223 UP000235965 UP000078200 UP000183832 UP000092461
Interpro
Gene 3D
ProteinModelPortal
A0A194QEL2
A0A0L7LBA3
A0A195FV70
A0A195B1Q4
E2BE86
F4WXX7
+ More
H9JES6 A0A067R7B8 A0A194QKC7 A0A2W1BR63 A0A2H1W476 A0A3S2LCA2 A0A0L7RGJ2 A0A212EJR9 A0A0C9RUC2 A0A3L8DY73 A0A026X3Q6 A0A151WTY0 A0A195CIJ3 A0A158NZ91 A0A0J9TUG9 K7IYV6 A0A1W4VFE8 A0A0Q9X0P2 A0A1B6EFX8 A0A1B6F054 A0A0J9TUG3 A0A0Q5WNY3 A0A1B6LBI4 A0A1B6DCF0 A0A0L0CE53 A0A232ERZ5 A0A1Y1L3H5 A0A1Y1KXP6 W8AVA6 W8AR81 B4P107 A0A1W4VE31 A0A1W4V1T0 B4HQB3 A0A0C9R9T6 B4J6K1 A0A139WMR8 A0A034VR99 A0A034VUQ8 A0A0M4E6M5 A0A139WM79 A0A1Y1L3G5 A0A0T6B0M2 E2AH21 A0A1Y1L4Y4 A0A2R7VRI3 A0A1Y1KXN3 A0A1Y1L045 A0A0J9TUH8 B3MIN9 Q9V9A9 A0A0J9R6U2 E1JGY6 A0A1I8P428 A0A1I8P422 A0A1I8P3Y7 A0A1W4VEI9 A0A1I8P433 B3NB27 A0A1W4VE25 B4LK14 A0A1W4V1S5 A0A1W4VFE3 B4MY49 A0A0Q9X1D6 A0A146M1J4 A0A0C9RFW1 A0A1I8M374 A0A1I8M384 A0A1I8M378 A0A1I8M379 A0A1W4WET8 A0A139WM70 A0A1Y1L171 A0A1W4W2X2 A0A034VRA7 A0A2J7PVL4 A0A1Y1L2F8 A0A034VUR2 A0A1A9UIW9 A0A1J1I7I3 A0A1J1IBB7 A0A0J9R6J4 A0A0J9R6I3 Q0E9N2 A0A1I8P3Y0 A0A0J9R6T3 A0A1I8P423 A0A1W4V279 U5EMW7 A0A1B0EXM3 A0A2H8TVP0 A0A1I8M376 A0A2J7PVK4
H9JES6 A0A067R7B8 A0A194QKC7 A0A2W1BR63 A0A2H1W476 A0A3S2LCA2 A0A0L7RGJ2 A0A212EJR9 A0A0C9RUC2 A0A3L8DY73 A0A026X3Q6 A0A151WTY0 A0A195CIJ3 A0A158NZ91 A0A0J9TUG9 K7IYV6 A0A1W4VFE8 A0A0Q9X0P2 A0A1B6EFX8 A0A1B6F054 A0A0J9TUG3 A0A0Q5WNY3 A0A1B6LBI4 A0A1B6DCF0 A0A0L0CE53 A0A232ERZ5 A0A1Y1L3H5 A0A1Y1KXP6 W8AVA6 W8AR81 B4P107 A0A1W4VE31 A0A1W4V1T0 B4HQB3 A0A0C9R9T6 B4J6K1 A0A139WMR8 A0A034VR99 A0A034VUQ8 A0A0M4E6M5 A0A139WM79 A0A1Y1L3G5 A0A0T6B0M2 E2AH21 A0A1Y1L4Y4 A0A2R7VRI3 A0A1Y1KXN3 A0A1Y1L045 A0A0J9TUH8 B3MIN9 Q9V9A9 A0A0J9R6U2 E1JGY6 A0A1I8P428 A0A1I8P422 A0A1I8P3Y7 A0A1W4VEI9 A0A1I8P433 B3NB27 A0A1W4VE25 B4LK14 A0A1W4V1S5 A0A1W4VFE3 B4MY49 A0A0Q9X1D6 A0A146M1J4 A0A0C9RFW1 A0A1I8M374 A0A1I8M384 A0A1I8M378 A0A1I8M379 A0A1W4WET8 A0A139WM70 A0A1Y1L171 A0A1W4W2X2 A0A034VRA7 A0A2J7PVL4 A0A1Y1L2F8 A0A034VUR2 A0A1A9UIW9 A0A1J1I7I3 A0A1J1IBB7 A0A0J9R6J4 A0A0J9R6I3 Q0E9N2 A0A1I8P3Y0 A0A0J9R6T3 A0A1I8P423 A0A1W4V279 U5EMW7 A0A1B0EXM3 A0A2H8TVP0 A0A1I8M376 A0A2J7PVK4
PDB
4EL1
E-value=2.78797e-06,
Score=127
Ontologies
GO
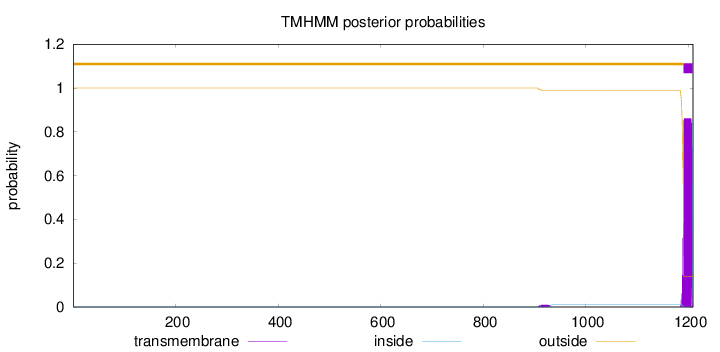
Topology
Subcellular location
Mitochondrion
Membrane
Membrane
Length:
1209
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
16.49972
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00002
outside
1 - 1190
TMhelix
1191 - 1208
inside
1209 - 1209
Population Genetic Test Statistics
Pi
208.505855
Theta
191.863857
Tajima's D
0.621169
CLR
0.145395
CSRT
0.551722413879306
Interpretation
Uncertain
