Pre Gene Modal
BGIBMGA008023
Annotation
PREDICTED:_uncharacterized_protein_LOC101735409_[Bombyx_mori]
Full name
Kynurenine 3-monooxygenase
Alternative Name
Kynurenine 3-hydroxylase
Location in the cell
Cytoplasmic Reliability : 3.327
Sequence
CDS
ATGCACCATCTGTCATCTGTCAGTAATGACTGCAAGCAATGCCCTAGAGTGCTGGCCGAGATAGAGCATATCGACGACGACGCTGACGCTGCAGGAATCAACTTTGTGAAGATCGACGACAGACAAATGGCCAAAGAGCACGGCGTGTTCGCGCTGCCGGCCGTCCTGTTCTTCAAGCTGGGCTCCAAGGATCCTGTTATATACGCCGGTGACCTATACGACGAGCAGCAACTGTTGAGTTGGTTGCTGACCCAAAAAAATCCAGCCGGAGACGTGATCGAGGCTCTCGAAGGCCAAGAGCTACTCGATTTGATACAAGAGTCAACATCATTGGCGGTTTACTTCTATCTGGAACTTCCAGACGGTCCTGACTGCGAGCAGTGCGCCGGCATCTTGGAGGAGCTGGAGAACATCGATGACGACTGCGACAGACACGGCATTAAGTTCGTTAAGACCCAAGATTATTCCATAGCCGAGTCATACGGCGTAACAGATTTTCCAGTATTGGTCTACTTCGAAAACAACATACCTAATGTGTACGAAGGATCTCTGACTGAGGAAGAGGAAGTTTTACAATGGCTGATTACACAGAAGACTGAAGATCGTATTGAACTTATTACTAGGGTGATGCTGGAAAAAATGGTTGAAGAAACGCAATATCTGGCCGTATACTTTTACAAAATTAATTGCCACATATGTGAACACATACTCGAAGGTTTGGAGAGCATTGATGATGAATGTGACGTGTACGGGATACATATGGTCAAGATCCAAGATCCGCAACTAGCCAAGCGATACTCCATAAAAACTTTCCCTGCTATGGTCTATTTCAGGAACGGCAATCCGTTGCTGTTTGAAGGTGATCTACAGAACGAAGAGTCTATTTTGGAGTGGCTCATCGATGACGACAATCGGGAACTAGCTGATGAAATTGAGTCCGTCAATGATCGCATGCTGGAGAGGCTGCTGTACGAGTCACACTTGCTCGCTGTGTTCTTCTATGACGATGAAGATTGTCCCGAATGTCACGAGATCCTAGAGGCTCTGGAGCAGATAGACGGCGAGGTGGACCAGTACGGCATCGACTTCGTGAAGATCGCCAGCCCGGAGGCCGCGGCCGCCCACAACATCATCAACATACCCTCGCTCGTGTACTTCAGGAAGCAGGTGCCGATGCTGTACGACGGCGATCTGCACCAGGTCGACAAGGTCCTGCAGTGGCTCACTTCCCAGGACGTCTTCGAGATTAAGAACGAAATCGAAGAAGTCAACAGGAAAATGTTAGACAAACTCTTAGAGGAGAACGAGTTCTTGGCGGTTTATTTCTATGAGAAGTCAACAGAAAGTCGCGCGGTTCTAGACAAGCTGGAGAACATTGACAGCGAAACTGATAACTTGGACATAACCTTCGTTAAAATGCAAGACCCGAGATACGCTCGGAAGTGGGGGGTGACTAAACTGCCAGCCATAGTCTACTTCAGGAAGAGATTCCCCAGCATCTACCGAGGTGACCTCATGTCAGAGGAGGAGGTCCTGGAATGGCTGCGGAAGAACAGGTTCCGTCAGCCCGAGCTGAACATATTCATGTACGCGCTGCTGGCGCTCTCCGTGGCCTTCATCATGTACACCGCCTTCCTGCTGCAATGCTTCAAGGCGACGCCTCCAGCTCAAACACAGCACCCGAAACAAGCGTGA
Protein
MHHLSSVSNDCKQCPRVLAEIEHIDDDADAAGINFVKIDDRQMAKEHGVFALPAVLFFKLGSKDPVIYAGDLYDEQQLLSWLLTQKNPAGDVIEALEGQELLDLIQESTSLAVYFYLELPDGPDCEQCAGILEELENIDDDCDRHGIKFVKTQDYSIAESYGVTDFPVLVYFENNIPNVYEGSLTEEEEVLQWLITQKTEDRIELITRVMLEKMVEETQYLAVYFYKINCHICEHILEGLESIDDECDVYGIHMVKIQDPQLAKRYSIKTFPAMVYFRNGNPLLFEGDLQNEESILEWLIDDDNRELADEIESVNDRMLERLLYESHLLAVFFYDDEDCPECHEILEALEQIDGEVDQYGIDFVKIASPEAAAAHNIINIPSLVYFRKQVPMLYDGDLHQVDKVLQWLTSQDVFEIKNEIEEVNRKMLDKLLEENEFLAVYFYEKSTESRAVLDKLENIDSETDNLDITFVKMQDPRYARKWGVTKLPAIVYFRKRFPSIYRGDLMSEEEVLEWLRKNRFRQPELNIFMYALLALSVAFIMYTAFLLQCFKATPPAQTQHPKQA
Summary
Description
Catalyzes the hydroxylation of L-kynurenine (L-Kyn) to form 3-hydroxy-L-kynurenine (L-3OHKyn). Required for synthesis of quinolinic acid.
Catalytic Activity
H(+) + L-kynurenine + NADPH + O2 = 3-hydroxy-L-kynurenine + H2O + NADP(+)
Cofactor
FAD
Similarity
Belongs to the aromatic-ring hydroxylase family. KMO subfamily.
Uniprot
A0A2W1BR63
A0A194QEL2
A0A2A4ITE2
A0A194QKC7
A0A212EJR9
A0A2H1W476
+ More
A0A3S2LCA2 H9JES6 K7IYV6 A0A088AN82 A0A026X3Q6 A0A0L7RGX5 F4WXX7 A0A195B1Q4 E2BE86 A0A1L8DTU9 A0A1L8DTZ5 E2AH19 A0A195FV70 A0A3L8DY73 A0A195CIJ3 A0A182JI66 A0A139WMQ6 A0A1Y1L4X3 Q7Q5G3 A0A067R7B8 A0A0M4E6M5 Q16RV7 A0A0Q9X1D6 A0A0J9R6T3 A0A1I8M379 A0A0J9TUH8 Q9V9A9 A0A0J9R7Y1 T1PDM0 A0A1I8M373 E1JGY5 A0A0J9R7X6 Q0E9N3 A0A1W4VFE3 A0A1W4VEJ2 A0A2J7PVK7 A0A2J7PVM0 A0A1Y1KXN7 A0A3B0JPC9 B4GC84 B4QDC0 A0A023EWC9 A0A1I8P3Y4 A0A1I8P428 A0A182LKY6 A0A182X900 A0A034VVF4 A0A2J7PVN9 A0A182UEC6 A0A034VUQ8 A0A034VUR2 A0A182UQN0 A0A0A1WLE2 B0XCN5 A0A151WUI2 A0A1B0DF10 A0A1W4W2X2 B0XLF6 A0A0J9R6I3 Q0E9N2 A0A1W4V279 A0A0A9YDH4 A0A1W4VEI9 A0A182PB84 A0A1W4WET8 A0A146M1J4 E0VU05 N6U1F2 A0A2S2QPU4 A0A182FF95 A0A2S2PLL5 A0A084VAX6 A0A182GP34 A0A1B0EXM3 A0A1J1IBB7 B4P107 A0A2S2N975 A0A1S4G7U5 A0A0P5TIE3 A0A0N8E2H2 A0A0P4YH92 A0A0N8CQG6 A0A0P5KYF6 A0A0P5ZHF9 A0A0P5EXE7 A0A0P5SS90 A0A0P5QUZ3 A0A0P6JK16 A0A0P5VWF6 A0A0P4ZT75 A0A0P5XAV2 A0A0N8D3Y4 A0A0N7ZJF2 A0A0P4XUL6
A0A3S2LCA2 H9JES6 K7IYV6 A0A088AN82 A0A026X3Q6 A0A0L7RGX5 F4WXX7 A0A195B1Q4 E2BE86 A0A1L8DTU9 A0A1L8DTZ5 E2AH19 A0A195FV70 A0A3L8DY73 A0A195CIJ3 A0A182JI66 A0A139WMQ6 A0A1Y1L4X3 Q7Q5G3 A0A067R7B8 A0A0M4E6M5 Q16RV7 A0A0Q9X1D6 A0A0J9R6T3 A0A1I8M379 A0A0J9TUH8 Q9V9A9 A0A0J9R7Y1 T1PDM0 A0A1I8M373 E1JGY5 A0A0J9R7X6 Q0E9N3 A0A1W4VFE3 A0A1W4VEJ2 A0A2J7PVK7 A0A2J7PVM0 A0A1Y1KXN7 A0A3B0JPC9 B4GC84 B4QDC0 A0A023EWC9 A0A1I8P3Y4 A0A1I8P428 A0A182LKY6 A0A182X900 A0A034VVF4 A0A2J7PVN9 A0A182UEC6 A0A034VUQ8 A0A034VUR2 A0A182UQN0 A0A0A1WLE2 B0XCN5 A0A151WUI2 A0A1B0DF10 A0A1W4W2X2 B0XLF6 A0A0J9R6I3 Q0E9N2 A0A1W4V279 A0A0A9YDH4 A0A1W4VEI9 A0A182PB84 A0A1W4WET8 A0A146M1J4 E0VU05 N6U1F2 A0A2S2QPU4 A0A182FF95 A0A2S2PLL5 A0A084VAX6 A0A182GP34 A0A1B0EXM3 A0A1J1IBB7 B4P107 A0A2S2N975 A0A1S4G7U5 A0A0P5TIE3 A0A0N8E2H2 A0A0P4YH92 A0A0N8CQG6 A0A0P5KYF6 A0A0P5ZHF9 A0A0P5EXE7 A0A0P5SS90 A0A0P5QUZ3 A0A0P6JK16 A0A0P5VWF6 A0A0P4ZT75 A0A0P5XAV2 A0A0N8D3Y4 A0A0N7ZJF2 A0A0P4XUL6
EC Number
1.14.13.9
Pubmed
28756777
26354079
22118469
19121390
20075255
24508170
+ More
21719571 20798317 30249741 18362917 19820115 28004739 12364791 14747013 17210077 24845553 17510324 17994087 18057021 22936249 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 20966253 25348373 25830018 25401762 26823975 20566863 23537049 24438588 26483478 17550304
21719571 20798317 30249741 18362917 19820115 28004739 12364791 14747013 17210077 24845553 17510324 17994087 18057021 22936249 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 20966253 25348373 25830018 25401762 26823975 20566863 23537049 24438588 26483478 17550304
EMBL
KZ149969
PZC76105.1
KQ459185
KPJ03385.1
NWSH01007220
PCG63035.1
+ More
KQ461198 KPJ05987.1 AGBW02014406 OWR41726.1 ODYU01006240 SOQ47899.1 RSAL01000048 RVE50479.1 BABH01023357 BABH01023358 BABH01023359 BABH01023360 BABH01023361 KK107013 EZA62912.1 KQ414596 KOC69941.1 GL888434 EGI60913.1 KQ976681 KYM78212.1 GL447768 EFN85931.1 GFDF01004277 JAV09807.1 GFDF01004207 JAV09877.1 GL439460 EFN67266.1 KQ981276 KYN43764.1 QOIP01000003 RLU24899.1 KQ977696 KYN00550.1 KQ971312 KYB29136.1 GEZM01071025 GEZM01071024 GEZM01071022 GEZM01071021 GEZM01071015 GEZM01071014 GEZM01071012 JAV66127.1 AAAB01008960 EAA11892.2 KK852827 KDR15395.1 CP012524 ALC42178.1 CH477696 EAT37127.1 CH963894 KRF98564.1 KRF98565.1 KRF98567.1 CM002911 KMY91798.1 KMY91795.1 AF216973 AE013599 AAF34746.1 AAN16126.1 KMY91789.1 KA646882 AFP61511.1 ACZ94346.1 KMY91784.1 ABI31025.2 NEVH01020956 PNF20361.1 PNF20371.1 GEZM01071026 GEZM01071023 GEZM01071020 GEZM01071019 GEZM01071018 GEZM01071017 GEZM01071016 GEZM01071013 JAV66132.1 OUUW01000001 SPP73028.1 CH479181 EDW31402.1 CM000362 EDX05907.1 GAPW01000053 JAC13545.1 GAKP01013137 JAC45815.1 PNF20370.1 GAKP01013125 GAKP01013121 JAC45827.1 GAKP01013131 GAKP01013120 JAC45832.1 GBXI01014610 JAC99681.1 DS232707 EDS44931.1 KQ982748 KYQ51295.1 AJVK01058476 DS234457 EDS34063.1 KMY91787.1 ABI31026.3 GBHO01014458 GBHO01014456 GBRD01003454 JAG29146.1 JAG29148.1 JAG62367.1 GDHC01004946 JAQ13683.1 DS235777 EEB16861.1 APGK01053421 APGK01053422 APGK01053423 APGK01053424 KB741223 ENN72382.1 GGMS01010544 MBY79747.1 GGMR01017721 MBY30340.1 ATLV01004734 ATLV01004735 ATLV01004736 ATLV01004737 ATLV01004738 ATLV01004739 ATLV01004740 KE524237 KFB35120.1 JXUM01014797 JXUM01014798 JXUM01014799 KQ560413 KXJ82542.1 AJWK01034186 CVRI01000043 CRK96270.1 CM000157 EDW89081.2 GGMR01001165 MBY13784.1 GDIP01125817 JAL77897.1 GDIQ01068292 JAN26445.1 GDIP01228660 JAI94741.1 GDIP01108919 JAL94795.1 GDIQ01182452 JAK69273.1 GDIP01045285 JAM58430.1 GDIQ01265617 JAJ86107.1 GDIP01141364 JAL62350.1 GDIQ01129525 JAL22201.1 GDIQ01004918 JAN89819.1 GDIP01108918 JAL94796.1 GDIP01209908 JAJ13494.1 GDIP01076371 JAM27344.1 GDIP01071175 JAM32540.1 GDIP01252160 GDIP01239521 GDIP01230262 GDIP01222615 GDIP01186762 GDIP01186761 GDIP01167061 GDIP01163062 GDIP01146768 GDIP01124681 GDIP01095329 GDIP01088073 GDIP01087073 GDIP01074885 GDIP01056078 GDIP01048337 GDIP01034884 JAI83880.1 GDIP01236506 JAI86895.1
KQ461198 KPJ05987.1 AGBW02014406 OWR41726.1 ODYU01006240 SOQ47899.1 RSAL01000048 RVE50479.1 BABH01023357 BABH01023358 BABH01023359 BABH01023360 BABH01023361 KK107013 EZA62912.1 KQ414596 KOC69941.1 GL888434 EGI60913.1 KQ976681 KYM78212.1 GL447768 EFN85931.1 GFDF01004277 JAV09807.1 GFDF01004207 JAV09877.1 GL439460 EFN67266.1 KQ981276 KYN43764.1 QOIP01000003 RLU24899.1 KQ977696 KYN00550.1 KQ971312 KYB29136.1 GEZM01071025 GEZM01071024 GEZM01071022 GEZM01071021 GEZM01071015 GEZM01071014 GEZM01071012 JAV66127.1 AAAB01008960 EAA11892.2 KK852827 KDR15395.1 CP012524 ALC42178.1 CH477696 EAT37127.1 CH963894 KRF98564.1 KRF98565.1 KRF98567.1 CM002911 KMY91798.1 KMY91795.1 AF216973 AE013599 AAF34746.1 AAN16126.1 KMY91789.1 KA646882 AFP61511.1 ACZ94346.1 KMY91784.1 ABI31025.2 NEVH01020956 PNF20361.1 PNF20371.1 GEZM01071026 GEZM01071023 GEZM01071020 GEZM01071019 GEZM01071018 GEZM01071017 GEZM01071016 GEZM01071013 JAV66132.1 OUUW01000001 SPP73028.1 CH479181 EDW31402.1 CM000362 EDX05907.1 GAPW01000053 JAC13545.1 GAKP01013137 JAC45815.1 PNF20370.1 GAKP01013125 GAKP01013121 JAC45827.1 GAKP01013131 GAKP01013120 JAC45832.1 GBXI01014610 JAC99681.1 DS232707 EDS44931.1 KQ982748 KYQ51295.1 AJVK01058476 DS234457 EDS34063.1 KMY91787.1 ABI31026.3 GBHO01014458 GBHO01014456 GBRD01003454 JAG29146.1 JAG29148.1 JAG62367.1 GDHC01004946 JAQ13683.1 DS235777 EEB16861.1 APGK01053421 APGK01053422 APGK01053423 APGK01053424 KB741223 ENN72382.1 GGMS01010544 MBY79747.1 GGMR01017721 MBY30340.1 ATLV01004734 ATLV01004735 ATLV01004736 ATLV01004737 ATLV01004738 ATLV01004739 ATLV01004740 KE524237 KFB35120.1 JXUM01014797 JXUM01014798 JXUM01014799 KQ560413 KXJ82542.1 AJWK01034186 CVRI01000043 CRK96270.1 CM000157 EDW89081.2 GGMR01001165 MBY13784.1 GDIP01125817 JAL77897.1 GDIQ01068292 JAN26445.1 GDIP01228660 JAI94741.1 GDIP01108919 JAL94795.1 GDIQ01182452 JAK69273.1 GDIP01045285 JAM58430.1 GDIQ01265617 JAJ86107.1 GDIP01141364 JAL62350.1 GDIQ01129525 JAL22201.1 GDIQ01004918 JAN89819.1 GDIP01108918 JAL94796.1 GDIP01209908 JAJ13494.1 GDIP01076371 JAM27344.1 GDIP01071175 JAM32540.1 GDIP01252160 GDIP01239521 GDIP01230262 GDIP01222615 GDIP01186762 GDIP01186761 GDIP01167061 GDIP01163062 GDIP01146768 GDIP01124681 GDIP01095329 GDIP01088073 GDIP01087073 GDIP01074885 GDIP01056078 GDIP01048337 GDIP01034884 JAI83880.1 GDIP01236506 JAI86895.1
Proteomes
UP000053268
UP000218220
UP000053240
UP000007151
UP000283053
UP000005204
+ More
UP000002358 UP000005203 UP000053097 UP000053825 UP000007755 UP000078540 UP000008237 UP000000311 UP000078541 UP000279307 UP000078542 UP000075880 UP000007266 UP000007062 UP000027135 UP000092553 UP000008820 UP000007798 UP000095301 UP000000803 UP000192221 UP000235965 UP000268350 UP000008744 UP000000304 UP000095300 UP000075882 UP000076407 UP000075902 UP000075903 UP000002320 UP000075809 UP000092462 UP000192223 UP000075885 UP000009046 UP000019118 UP000069272 UP000030765 UP000069940 UP000249989 UP000092461 UP000183832 UP000002282
UP000002358 UP000005203 UP000053097 UP000053825 UP000007755 UP000078540 UP000008237 UP000000311 UP000078541 UP000279307 UP000078542 UP000075880 UP000007266 UP000007062 UP000027135 UP000092553 UP000008820 UP000007798 UP000095301 UP000000803 UP000192221 UP000235965 UP000268350 UP000008744 UP000000304 UP000095300 UP000075882 UP000076407 UP000075902 UP000075903 UP000002320 UP000075809 UP000092462 UP000192223 UP000075885 UP000009046 UP000019118 UP000069272 UP000030765 UP000069940 UP000249989 UP000092461 UP000183832 UP000002282
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BR63
A0A194QEL2
A0A2A4ITE2
A0A194QKC7
A0A212EJR9
A0A2H1W476
+ More
A0A3S2LCA2 H9JES6 K7IYV6 A0A088AN82 A0A026X3Q6 A0A0L7RGX5 F4WXX7 A0A195B1Q4 E2BE86 A0A1L8DTU9 A0A1L8DTZ5 E2AH19 A0A195FV70 A0A3L8DY73 A0A195CIJ3 A0A182JI66 A0A139WMQ6 A0A1Y1L4X3 Q7Q5G3 A0A067R7B8 A0A0M4E6M5 Q16RV7 A0A0Q9X1D6 A0A0J9R6T3 A0A1I8M379 A0A0J9TUH8 Q9V9A9 A0A0J9R7Y1 T1PDM0 A0A1I8M373 E1JGY5 A0A0J9R7X6 Q0E9N3 A0A1W4VFE3 A0A1W4VEJ2 A0A2J7PVK7 A0A2J7PVM0 A0A1Y1KXN7 A0A3B0JPC9 B4GC84 B4QDC0 A0A023EWC9 A0A1I8P3Y4 A0A1I8P428 A0A182LKY6 A0A182X900 A0A034VVF4 A0A2J7PVN9 A0A182UEC6 A0A034VUQ8 A0A034VUR2 A0A182UQN0 A0A0A1WLE2 B0XCN5 A0A151WUI2 A0A1B0DF10 A0A1W4W2X2 B0XLF6 A0A0J9R6I3 Q0E9N2 A0A1W4V279 A0A0A9YDH4 A0A1W4VEI9 A0A182PB84 A0A1W4WET8 A0A146M1J4 E0VU05 N6U1F2 A0A2S2QPU4 A0A182FF95 A0A2S2PLL5 A0A084VAX6 A0A182GP34 A0A1B0EXM3 A0A1J1IBB7 B4P107 A0A2S2N975 A0A1S4G7U5 A0A0P5TIE3 A0A0N8E2H2 A0A0P4YH92 A0A0N8CQG6 A0A0P5KYF6 A0A0P5ZHF9 A0A0P5EXE7 A0A0P5SS90 A0A0P5QUZ3 A0A0P6JK16 A0A0P5VWF6 A0A0P4ZT75 A0A0P5XAV2 A0A0N8D3Y4 A0A0N7ZJF2 A0A0P4XUL6
A0A3S2LCA2 H9JES6 K7IYV6 A0A088AN82 A0A026X3Q6 A0A0L7RGX5 F4WXX7 A0A195B1Q4 E2BE86 A0A1L8DTU9 A0A1L8DTZ5 E2AH19 A0A195FV70 A0A3L8DY73 A0A195CIJ3 A0A182JI66 A0A139WMQ6 A0A1Y1L4X3 Q7Q5G3 A0A067R7B8 A0A0M4E6M5 Q16RV7 A0A0Q9X1D6 A0A0J9R6T3 A0A1I8M379 A0A0J9TUH8 Q9V9A9 A0A0J9R7Y1 T1PDM0 A0A1I8M373 E1JGY5 A0A0J9R7X6 Q0E9N3 A0A1W4VFE3 A0A1W4VEJ2 A0A2J7PVK7 A0A2J7PVM0 A0A1Y1KXN7 A0A3B0JPC9 B4GC84 B4QDC0 A0A023EWC9 A0A1I8P3Y4 A0A1I8P428 A0A182LKY6 A0A182X900 A0A034VVF4 A0A2J7PVN9 A0A182UEC6 A0A034VUQ8 A0A034VUR2 A0A182UQN0 A0A0A1WLE2 B0XCN5 A0A151WUI2 A0A1B0DF10 A0A1W4W2X2 B0XLF6 A0A0J9R6I3 Q0E9N2 A0A1W4V279 A0A0A9YDH4 A0A1W4VEI9 A0A182PB84 A0A1W4WET8 A0A146M1J4 E0VU05 N6U1F2 A0A2S2QPU4 A0A182FF95 A0A2S2PLL5 A0A084VAX6 A0A182GP34 A0A1B0EXM3 A0A1J1IBB7 B4P107 A0A2S2N975 A0A1S4G7U5 A0A0P5TIE3 A0A0N8E2H2 A0A0P4YH92 A0A0N8CQG6 A0A0P5KYF6 A0A0P5ZHF9 A0A0P5EXE7 A0A0P5SS90 A0A0P5QUZ3 A0A0P6JK16 A0A0P5VWF6 A0A0P4ZT75 A0A0P5XAV2 A0A0N8D3Y4 A0A0N7ZJF2 A0A0P4XUL6
PDB
6ESX
E-value=0.000192526,
Score=108
Ontologies
GO
Topology
Subcellular location
Mitochondrion
Membrane
Membrane
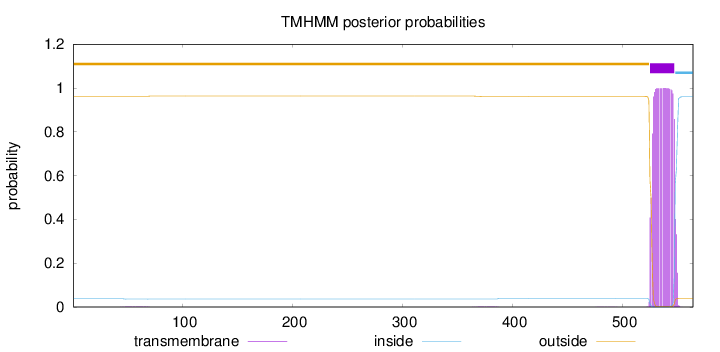
Length:
564
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.45193
Exp number, first 60 AAs:
0.02453
Total prob of N-in:
0.03747
outside
1 - 524
TMhelix
525 - 547
inside
548 - 564
Population Genetic Test Statistics
Pi
232.238187
Theta
174.452729
Tajima's D
0.896891
CLR
0.935211
CSRT
0.630018499075046
Interpretation
Uncertain
