Pre Gene Modal
BGIBMGA008049
Annotation
monoacylglycerol_acyl_transferase_[Manduca_sexta]
Location in the cell
Cytoplasmic Reliability : 1.266 Mitochondrial Reliability : 1.875
Sequence
CDS
ATGGTAGGAGGAGCCGCTGAGGCGCTTGACTCGCATCCTGGAAAGTACAAAATCATTCTCAGCCGACGAAAGGGATTTATTCGCATCGCTATGAAGTCAGGAGCGTCCCTTGTACCCGTTTTTTCGTTTGGTGAAACTGACGTTTTCCGACCTCTCAATAACCCCGAGAACAGCCTGCTGAGGAGGATACAAGAAAAAGTGCGTCAGATAACTGGAGTTTCACCAGTCTTCCCTCTTGGGAGGGGCATGTTCCAGTATTCGTTCGGGGTTGTACCTCTCAGGAGTCCCGTTACAACTGTTGTGGGTGCTCCGATGGAGGTGACGAGGAACCTGGAGCCCACAGACGAGGAAGTGGATGAGGTCCACGCGGAGTTCACGAAGCGACTCGTCCAGCTCTTCGAGAACGAGAAGTCCAAGTACTTGAAGAATCACGAGGAAGTGCAGCTTGTCATCACCTAG
Protein
MVGGAAEALDSHPGKYKIILSRRKGFIRIAMKSGASLVPVFSFGETDVFRPLNNPENSLLRRIQEKVRQITGVSPVFPLGRGMFQYSFGVVPLRSPVTTVVGAPMEVTRNLEPTDEEVDEVHAEFTKRLVQLFENEKSKYLKNHEEVQLVIT
Summary
Uniprot
A0A0G3EZA8
H9JEV2
A0A023J8Y6
A0A023J967
A0A212EJT6
H9JEV1
+ More
A0A2A4IU63 A0A291P132 A0A0P0DZ59 A0A291P136 A0A2W1BR28 S4P574 A0A1V0M8B2 A0A2H1W4C9 A0A088MCZ8 A0A194QIV6 A0A194QK89 A0A1E1VZU1 A0A0L7LB93 A0A232ES18 K7IYV8 A0A2J7PVK2 A0A2A3EBP2 A0A088AT65 A0A0T6B0G8 A0A195FTL3 A0A158NYM6 A0A1Y1M1N3 A0A195DE24 A0A195CIK5 A0A026X5Z1 B4J4Z0 A0A151WTY2 Q7K1C2 A0A1B0DR23 E9IUY0 F4WXX9 B4LMJ2 A0A1Y1NLC3 A0A0M4E837 A0A0K8UDQ8 A0A1L8DYI7 A0A195CII8 A0A0A1WH54 E2BE83 A0A1L8DY79 A0A154PDD6 W8ASZ4 B4HR66 A0A0C9Q1U7 A0A3Q0IKS5 A0A1B0GL48 A0A0K8V4G6 A0A1D2NKJ3 B4QER8 A0A1S3KAF8 A0A1Y1MYE2 B3MHT8 A0A034WH55 A0A0L0C2V4 A0A151IKB8 A0A0Q9X7D1 A0A0L7RG55 B4KSX6 B4P228 A0A1B6J4Y1 A0A1J1IUY8 B4MPK0 E2AAY9 N6TVA7 A0A3B0J0T3 A0A1B6MBW2 A0A1W4VYU1 A0A182GP38 B3N9U5 A0A3B0JM49 A0A1S3JYS2 A0A182ILZ7 A0A1A9WJZ7 U4UAX9 B5E0G7 B4GBF0 J3JWV0 A0A195CJC8 A0A182WKQ3 A0A0A9X5J6 A0A091NY11 W5JD56 A0A016WDJ9 A0A0A9XCV1 A0A084WAA2 A0A182Q0W4 B3MHT7 G5AS70 A0A147BP68 A0A131XXT7 A0A195CJD4 A0A091URJ0 A0A3M0KG59 A0A016WBS5
A0A2A4IU63 A0A291P132 A0A0P0DZ59 A0A291P136 A0A2W1BR28 S4P574 A0A1V0M8B2 A0A2H1W4C9 A0A088MCZ8 A0A194QIV6 A0A194QK89 A0A1E1VZU1 A0A0L7LB93 A0A232ES18 K7IYV8 A0A2J7PVK2 A0A2A3EBP2 A0A088AT65 A0A0T6B0G8 A0A195FTL3 A0A158NYM6 A0A1Y1M1N3 A0A195DE24 A0A195CIK5 A0A026X5Z1 B4J4Z0 A0A151WTY2 Q7K1C2 A0A1B0DR23 E9IUY0 F4WXX9 B4LMJ2 A0A1Y1NLC3 A0A0M4E837 A0A0K8UDQ8 A0A1L8DYI7 A0A195CII8 A0A0A1WH54 E2BE83 A0A1L8DY79 A0A154PDD6 W8ASZ4 B4HR66 A0A0C9Q1U7 A0A3Q0IKS5 A0A1B0GL48 A0A0K8V4G6 A0A1D2NKJ3 B4QER8 A0A1S3KAF8 A0A1Y1MYE2 B3MHT8 A0A034WH55 A0A0L0C2V4 A0A151IKB8 A0A0Q9X7D1 A0A0L7RG55 B4KSX6 B4P228 A0A1B6J4Y1 A0A1J1IUY8 B4MPK0 E2AAY9 N6TVA7 A0A3B0J0T3 A0A1B6MBW2 A0A1W4VYU1 A0A182GP38 B3N9U5 A0A3B0JM49 A0A1S3JYS2 A0A182ILZ7 A0A1A9WJZ7 U4UAX9 B5E0G7 B4GBF0 J3JWV0 A0A195CJC8 A0A182WKQ3 A0A0A9X5J6 A0A091NY11 W5JD56 A0A016WDJ9 A0A0A9XCV1 A0A084WAA2 A0A182Q0W4 B3MHT7 G5AS70 A0A147BP68 A0A131XXT7 A0A195CJD4 A0A091URJ0 A0A3M0KG59 A0A016WBS5
Pubmed
19121390
25263765
22118469
28986331
26445454
28756777
+ More
23622113 28349297 26385554 26354079 26227816 28648823 20075255 21347285 28004739 24508170 30249741 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 21719571 18057021 25830018 20798317 24495485 27289101 22936249 25348373 26108605 17550304 23537049 26483478 15632085 22516182 25401762 26823975 20920257 23761445 25730766 24438588 21993625 29652888
23622113 28349297 26385554 26354079 26227816 28648823 20075255 21347285 28004739 24508170 30249741 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 21719571 18057021 25830018 20798317 24495485 27289101 22936249 25348373 26108605 17550304 23537049 26483478 15632085 22516182 25401762 26823975 20920257 23761445 25730766 24438588 21993625 29652888
EMBL
KR560051
AKJ70771.1
BABH01023356
BABH01023357
KF800700
AHH25136.1
+ More
KF800699 AHH25135.1 AGBW02014406 OWR41725.1 BABH01023355 NWSH01007220 PCG63036.1 MF687652 ATJ44578.1 KT261724 ALJ30264.1 MF706186 ATJ44613.1 KZ149969 PZC76104.1 GAIX01005704 JAA86856.1 KU755514 ARD71224.1 ODYU01006240 SOQ47898.1 KJ579217 AIN34693.1 KQ459185 KPJ03386.1 KQ461198 KPJ05988.1 GDQN01010799 JAT80255.1 JTDY01001873 KOB72665.1 NNAY01002528 OXU21132.1 NEVH01020956 PNF20359.1 KZ288292 PBC29138.1 LJIG01016421 KRT80651.1 KQ981276 KYN43766.1 ADTU01004076 ADTU01004077 GEZM01045470 GEZM01045469 JAV77796.1 KQ980953 KYN11143.1 KQ977696 KYN00548.1 KK107013 QOIP01000003 EZA62914.1 RLU24901.1 CH916367 EDW01696.1 KQ982748 KYQ51298.1 AE013599 AY069178 AAF59201.1 AAL39323.1 AJVK01019569 GL766096 EFZ15622.1 GL888434 EGI60915.1 CH940648 EDW59979.1 KRF79182.1 GEZM01000294 GEZM01000293 JAV98438.1 CP012524 ALC40808.1 GDHF01027522 JAI24792.1 GFDF01002682 JAV11402.1 KYN00545.1 GBXI01016492 GBXI01008971 JAC97799.1 JAD05321.1 GL447768 EFN85928.1 GFDF01002681 JAV11403.1 KQ434878 KZC09827.1 GAMC01018757 JAB87798.1 CH480816 EDW46809.1 GBYB01007947 GBYB01007948 JAG77714.1 JAG77715.1 AJWK01034186 GDHF01018824 GDHF01016125 GDHF01015434 GDHF01004697 GDHF01002420 JAI33490.1 JAI36189.1 JAI36880.1 JAI47617.1 JAI49894.1 LJIJ01000019 ODN05635.1 CM000362 CM002911 EDX06064.1 KMY92077.1 GEZM01017419 GEZM01017418 GEZM01017416 JAV90701.1 CH902619 EDV36925.2 GAKP01004928 JAC54024.1 JRES01000975 KNC26567.1 KQ977224 KYN04776.1 CH933808 KRG04140.1 KQ414596 KOC69937.1 EDW08473.1 KRG04139.1 CM000157 EDW89229.1 GECU01013481 JAS94225.1 CVRI01000059 CRL03396.1 CH963849 EDW74039.1 GL438222 EFN69400.1 APGK01053258 APGK01053259 KB741223 ENN72331.1 OUUW01000001 SPP74347.1 GEBQ01006608 JAT33369.1 JXUM01014810 KQ560413 KXJ82546.1 CH954177 EDV59641.1 SPP74346.1 KB632273 ERL91069.1 CM000071 EDY68989.1 CH479181 EDW32252.1 BT127718 AEE62680.1 KYN00542.1 GBHO01035686 GBHO01028673 GDHC01009211 JAG07918.1 JAG14931.1 JAQ09418.1 KK842088 KFP84360.1 ADMH02001858 ETN60830.1 KE125468 JARK01000431 EPB68262.1 EYC37043.1 GBHO01028674 GBHO01028671 JAG14930.1 JAG14933.1 ATLV01022056 KE525327 KFB47146.1 AXCN02000417 EDV36924.1 JH166727 GEBF01007595 EHA99876.1 JAN96037.1 GEGO01003122 JAR92282.1 GEFM01004299 JAP71497.1 KYN00547.1 KL409863 KFQ92575.1 QRBI01000107 RMC12035.1 EYC37045.1
KF800699 AHH25135.1 AGBW02014406 OWR41725.1 BABH01023355 NWSH01007220 PCG63036.1 MF687652 ATJ44578.1 KT261724 ALJ30264.1 MF706186 ATJ44613.1 KZ149969 PZC76104.1 GAIX01005704 JAA86856.1 KU755514 ARD71224.1 ODYU01006240 SOQ47898.1 KJ579217 AIN34693.1 KQ459185 KPJ03386.1 KQ461198 KPJ05988.1 GDQN01010799 JAT80255.1 JTDY01001873 KOB72665.1 NNAY01002528 OXU21132.1 NEVH01020956 PNF20359.1 KZ288292 PBC29138.1 LJIG01016421 KRT80651.1 KQ981276 KYN43766.1 ADTU01004076 ADTU01004077 GEZM01045470 GEZM01045469 JAV77796.1 KQ980953 KYN11143.1 KQ977696 KYN00548.1 KK107013 QOIP01000003 EZA62914.1 RLU24901.1 CH916367 EDW01696.1 KQ982748 KYQ51298.1 AE013599 AY069178 AAF59201.1 AAL39323.1 AJVK01019569 GL766096 EFZ15622.1 GL888434 EGI60915.1 CH940648 EDW59979.1 KRF79182.1 GEZM01000294 GEZM01000293 JAV98438.1 CP012524 ALC40808.1 GDHF01027522 JAI24792.1 GFDF01002682 JAV11402.1 KYN00545.1 GBXI01016492 GBXI01008971 JAC97799.1 JAD05321.1 GL447768 EFN85928.1 GFDF01002681 JAV11403.1 KQ434878 KZC09827.1 GAMC01018757 JAB87798.1 CH480816 EDW46809.1 GBYB01007947 GBYB01007948 JAG77714.1 JAG77715.1 AJWK01034186 GDHF01018824 GDHF01016125 GDHF01015434 GDHF01004697 GDHF01002420 JAI33490.1 JAI36189.1 JAI36880.1 JAI47617.1 JAI49894.1 LJIJ01000019 ODN05635.1 CM000362 CM002911 EDX06064.1 KMY92077.1 GEZM01017419 GEZM01017418 GEZM01017416 JAV90701.1 CH902619 EDV36925.2 GAKP01004928 JAC54024.1 JRES01000975 KNC26567.1 KQ977224 KYN04776.1 CH933808 KRG04140.1 KQ414596 KOC69937.1 EDW08473.1 KRG04139.1 CM000157 EDW89229.1 GECU01013481 JAS94225.1 CVRI01000059 CRL03396.1 CH963849 EDW74039.1 GL438222 EFN69400.1 APGK01053258 APGK01053259 KB741223 ENN72331.1 OUUW01000001 SPP74347.1 GEBQ01006608 JAT33369.1 JXUM01014810 KQ560413 KXJ82546.1 CH954177 EDV59641.1 SPP74346.1 KB632273 ERL91069.1 CM000071 EDY68989.1 CH479181 EDW32252.1 BT127718 AEE62680.1 KYN00542.1 GBHO01035686 GBHO01028673 GDHC01009211 JAG07918.1 JAG14931.1 JAQ09418.1 KK842088 KFP84360.1 ADMH02001858 ETN60830.1 KE125468 JARK01000431 EPB68262.1 EYC37043.1 GBHO01028674 GBHO01028671 JAG14930.1 JAG14933.1 ATLV01022056 KE525327 KFB47146.1 AXCN02000417 EDV36924.1 JH166727 GEBF01007595 EHA99876.1 JAN96037.1 GEGO01003122 JAR92282.1 GEFM01004299 JAP71497.1 KYN00547.1 KL409863 KFQ92575.1 QRBI01000107 RMC12035.1 EYC37045.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000037510
+ More
UP000215335 UP000002358 UP000235965 UP000242457 UP000005203 UP000078541 UP000005205 UP000078492 UP000078542 UP000053097 UP000279307 UP000001070 UP000075809 UP000000803 UP000092462 UP000007755 UP000008792 UP000092553 UP000008237 UP000076502 UP000001292 UP000079169 UP000092461 UP000094527 UP000000304 UP000085678 UP000007801 UP000037069 UP000009192 UP000053825 UP000002282 UP000183832 UP000007798 UP000000311 UP000019118 UP000268350 UP000192221 UP000069940 UP000249989 UP000008711 UP000075880 UP000091820 UP000030742 UP000001819 UP000008744 UP000075920 UP000000673 UP000024635 UP000030765 UP000075886 UP000006813 UP000053283 UP000269221
UP000215335 UP000002358 UP000235965 UP000242457 UP000005203 UP000078541 UP000005205 UP000078492 UP000078542 UP000053097 UP000279307 UP000001070 UP000075809 UP000000803 UP000092462 UP000007755 UP000008792 UP000092553 UP000008237 UP000076502 UP000001292 UP000079169 UP000092461 UP000094527 UP000000304 UP000085678 UP000007801 UP000037069 UP000009192 UP000053825 UP000002282 UP000183832 UP000007798 UP000000311 UP000019118 UP000268350 UP000192221 UP000069940 UP000249989 UP000008711 UP000075880 UP000091820 UP000030742 UP000001819 UP000008744 UP000075920 UP000000673 UP000024635 UP000030765 UP000075886 UP000006813 UP000053283 UP000269221
Pfam
PF03982 DAGAT
Interpro
IPR007130
DAGAT
ProteinModelPortal
A0A0G3EZA8
H9JEV2
A0A023J8Y6
A0A023J967
A0A212EJT6
H9JEV1
+ More
A0A2A4IU63 A0A291P132 A0A0P0DZ59 A0A291P136 A0A2W1BR28 S4P574 A0A1V0M8B2 A0A2H1W4C9 A0A088MCZ8 A0A194QIV6 A0A194QK89 A0A1E1VZU1 A0A0L7LB93 A0A232ES18 K7IYV8 A0A2J7PVK2 A0A2A3EBP2 A0A088AT65 A0A0T6B0G8 A0A195FTL3 A0A158NYM6 A0A1Y1M1N3 A0A195DE24 A0A195CIK5 A0A026X5Z1 B4J4Z0 A0A151WTY2 Q7K1C2 A0A1B0DR23 E9IUY0 F4WXX9 B4LMJ2 A0A1Y1NLC3 A0A0M4E837 A0A0K8UDQ8 A0A1L8DYI7 A0A195CII8 A0A0A1WH54 E2BE83 A0A1L8DY79 A0A154PDD6 W8ASZ4 B4HR66 A0A0C9Q1U7 A0A3Q0IKS5 A0A1B0GL48 A0A0K8V4G6 A0A1D2NKJ3 B4QER8 A0A1S3KAF8 A0A1Y1MYE2 B3MHT8 A0A034WH55 A0A0L0C2V4 A0A151IKB8 A0A0Q9X7D1 A0A0L7RG55 B4KSX6 B4P228 A0A1B6J4Y1 A0A1J1IUY8 B4MPK0 E2AAY9 N6TVA7 A0A3B0J0T3 A0A1B6MBW2 A0A1W4VYU1 A0A182GP38 B3N9U5 A0A3B0JM49 A0A1S3JYS2 A0A182ILZ7 A0A1A9WJZ7 U4UAX9 B5E0G7 B4GBF0 J3JWV0 A0A195CJC8 A0A182WKQ3 A0A0A9X5J6 A0A091NY11 W5JD56 A0A016WDJ9 A0A0A9XCV1 A0A084WAA2 A0A182Q0W4 B3MHT7 G5AS70 A0A147BP68 A0A131XXT7 A0A195CJD4 A0A091URJ0 A0A3M0KG59 A0A016WBS5
A0A2A4IU63 A0A291P132 A0A0P0DZ59 A0A291P136 A0A2W1BR28 S4P574 A0A1V0M8B2 A0A2H1W4C9 A0A088MCZ8 A0A194QIV6 A0A194QK89 A0A1E1VZU1 A0A0L7LB93 A0A232ES18 K7IYV8 A0A2J7PVK2 A0A2A3EBP2 A0A088AT65 A0A0T6B0G8 A0A195FTL3 A0A158NYM6 A0A1Y1M1N3 A0A195DE24 A0A195CIK5 A0A026X5Z1 B4J4Z0 A0A151WTY2 Q7K1C2 A0A1B0DR23 E9IUY0 F4WXX9 B4LMJ2 A0A1Y1NLC3 A0A0M4E837 A0A0K8UDQ8 A0A1L8DYI7 A0A195CII8 A0A0A1WH54 E2BE83 A0A1L8DY79 A0A154PDD6 W8ASZ4 B4HR66 A0A0C9Q1U7 A0A3Q0IKS5 A0A1B0GL48 A0A0K8V4G6 A0A1D2NKJ3 B4QER8 A0A1S3KAF8 A0A1Y1MYE2 B3MHT8 A0A034WH55 A0A0L0C2V4 A0A151IKB8 A0A0Q9X7D1 A0A0L7RG55 B4KSX6 B4P228 A0A1B6J4Y1 A0A1J1IUY8 B4MPK0 E2AAY9 N6TVA7 A0A3B0J0T3 A0A1B6MBW2 A0A1W4VYU1 A0A182GP38 B3N9U5 A0A3B0JM49 A0A1S3JYS2 A0A182ILZ7 A0A1A9WJZ7 U4UAX9 B5E0G7 B4GBF0 J3JWV0 A0A195CJC8 A0A182WKQ3 A0A0A9X5J6 A0A091NY11 W5JD56 A0A016WDJ9 A0A0A9XCV1 A0A084WAA2 A0A182Q0W4 B3MHT7 G5AS70 A0A147BP68 A0A131XXT7 A0A195CJD4 A0A091URJ0 A0A3M0KG59 A0A016WBS5
Ontologies
GO
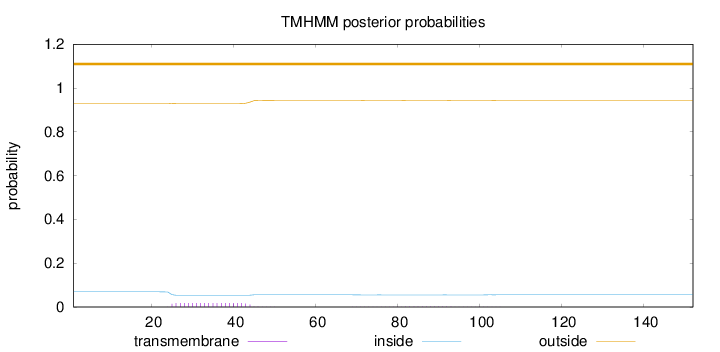
Topology
Length:
152
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.401
Exp number, first 60 AAs:
0.35719
Total prob of N-in:
0.06913
outside
1 - 152
Population Genetic Test Statistics
Pi
220.112501
Theta
159.779267
Tajima's D
0.831689
CLR
16.260811
CSRT
0.605919704014799
Interpretation
Uncertain
