Gene
KWMTBOMO04867
Pre Gene Modal
BGIBMGA008025
Annotation
PREDICTED:_set1/Ash2_histone_methyltransferase_complex_subunit_ASH2_[Papilio_xuthus]
Full name
Set1/Ash2 histone methyltransferase complex subunit ASH2
Alternative Name
Absent, small, or homeotic discs protein 2
Location in the cell
Mitochondrial Reliability : 1.905
Sequence
CDS
ATGGATAAAAATAAAGGAAATAAAACTTCTGCTGAATCTCAAGGGACATGCTACTGCGGTAAAGATCGTAATCTGAACATAGCTGAGTTATTATGTGCATCATGCAATAGATGGTTTCACGAATCCTGTATCGGTTATCAACTTGGCAAGCTGGTACCTTTCATGACAAATTATCTCTTTATTTGTAAGAACTGCTCTCCTACTGGTTTGGAAACATTCAAAAAAAATCAAGCACCATTCCCGCAAATGTGTCTAACTGCAATTGCTAATCTTCAACAAGATAGTGTGAAAAATGGTAGCAATAGACGTCTGTTCAGCAAAGACCGAGAGATCATACCATACATTGACCAGTATTGGGAAGCTATGACCACAATGCCCAGAAGGGTGACCCAGTCGTGGTATGCTACAGTACAGAGAGCTCTAATCAAGGATATCCAGGTTTTGTTCACATATGAAGAAGACTCGTCTGATGGTCCCATGTTTGGGCTCTGCAACGCAGAACTGACAATTATTAAACCTAACTACGAAGCCATGATCAAGCATGGTCAGCTCAAAGTCACAGATATGGGTATAGCTACAATTCCACTAGCCGGCAACGTGAAGGGCAGGCAAGGTAAGCGCAGGCCTGCGGTCGGCAGCGCGGCCGAGACCGGGGCTCCCATCGGCAAGAAAGGCCGTAACGCCGATGTCACCGCTCTGAAACTGCCCTCGCACGGCTACCCCACCGAGCATCCCTTCAACAAAGACGGTTACAGATACATCTTGGCGGAACCAGACCCGCACGCGCCCTTCAGACAGGAGTTCGACGAGAGCAACGAGTGGAGCGGCAAGCCGATCCCGGGCTGGCTGTACCGGCCGCTGTGCCCGGGCGGCGTGCTGCTGGCGCTGCACGACCGCGCGCCGCAGCTGCGCGTGGCCGAGGACCGGCTGGCCGTCACGGGCGACCGCGGCTACTGCACGGTGCGCGCCACGCACGGCGTGTCGCGCGGCAGCTGGTACTGGGAGGCGTGCGTGGAGGAGCTGCCGGAGGGCGCGGGGGTGCGCGCGGGGTGGGGCCGCCGCGTGGCCAACCTGCAGGCGCCGCTCGGGTACGACAAGTTCGGCTACTCGTGGCGGAGTCGCAAGGGGACCAGGTTCCACGAGTCCCGAGGCAAGCACTACAGCGGTGCGTACGGCGAGGGCGACACGCTGGGCTTCCTCATCGTGCTGCCGGACTGCGCCTCCACCAAGTTCACGCCCAACACCTACAAGGACCGGCCTCTAGTCAAATTCAAGAGCCACCTGTACTACGAGGACAAGGACAACATCCAGGAGTGCCTCAACAACCTGAAGCAGCTCCCCGGCAGCAAGATCATATTCTTCAAGAACGGCGAGTGCCAGGGCGAGGCCTTCAGTGACATCTACCGCGGCTGCTACTACCCCGCCGTCTCGCTGCACCGCACCGCCACCCTCAGCGTCAACTTCGGGCCCAGCTTCAAGTACCAGCCCTCCACGCACTACCCCTACAGACCTATGTCAGAAAAAGCGGAAGAAGCAATATGCGAACAAACGATGGCCGACATGCTGTATCTGACCGAGAACGAGGGCAAGCTGCGCCTGGACAACTTCAACATTTGA
Protein
MDKNKGNKTSAESQGTCYCGKDRNLNIAELLCASCNRWFHESCIGYQLGKLVPFMTNYLFICKNCSPTGLETFKKNQAPFPQMCLTAIANLQQDSVKNGSNRRLFSKDREIIPYIDQYWEAMTTMPRRVTQSWYATVQRALIKDIQVLFTYEEDSSDGPMFGLCNAELTIIKPNYEAMIKHGQLKVTDMGIATIPLAGNVKGRQGKRRPAVGSAAETGAPIGKKGRNADVTALKLPSHGYPTEHPFNKDGYRYILAEPDPHAPFRQEFDESNEWSGKPIPGWLYRPLCPGGVLLALHDRAPQLRVAEDRLAVTGDRGYCTVRATHGVSRGSWYWEACVEELPEGAGVRAGWGRRVANLQAPLGYDKFGYSWRSRKGTRFHESRGKHYSGAYGEGDTLGFLIVLPDCASTKFTPNTYKDRPLVKFKSHLYYEDKDNIQECLNNLKQLPGSKIIFFKNGECQGEAFSDIYRGCYYPAVSLHRTATLSVNFGPSFKYQPSTHYPYRPMSEKAEEAICEQTMADMLYLTENEGKLRLDNFNI
Summary
Description
Transcriptional regulator. Regulates a number of genes involved in wing development including activation of net and bs and repression of rho and kni and controls vein-intervein patterning during wing development. Required for correct expression of a number of homeotic genes including Scr in the first leg imaginal disk and Ubx in the third leg imaginal disk and haltere disks. Required for stabilization of the histone-lysine N-methyltransferase trr and for trimethylation of 'Lys-4' of histone H3. Plays a role in maintenance of transcriptionally active chromatin through down-regulation of histone H1 hyperphosphorylation.
Subunit
Core component of several methyltransferase-containing complexes. Component of the SET1 complex, composed at least of the catalytic subunit Set1, wds/WDR5, Wdr82, Rbbp5, ash2, Cfp1/CXXC1, hcf and Dpy-30L1. Component of the MLL3/4 complex composed at least of the catalytic subunit trr, ash2, Rbbp5, Dpy-30L1, wds, hcf, ptip, Pa1, Utx, Lpt and Ncoa6. Interacts with hcf, sktl and trr.
Keywords
Alternative splicing
Chromosome
Complete proteome
Developmental protein
DNA-binding
Metal-binding
Nucleus
Reference proteome
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Set1/Ash2 histone methyltransferase complex subunit ASH2
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JES8
A0A2H1W9Q6
A0A194QCW9
A0A212EJS5
A0A1Y1MNB3
D6X3Q8
+ More
E9J271 A0A151I3D9 A0A151IW66 E2ABY2 A0A026X3K3 A0A151WIR0 F4WMV2 A0A195FV15 A0A2J7QW12 A0A0N0BGY4 A0A067R8L5 A0A1B6ERW4 A0A0C9RK78 A0A0C9RK43 A0A088A739 A0A2A3E6E0 A0A0L0C6Q0 A0A1B6LI47 A0A154PC03 A0A347ZJ80 E2BJ62 A0A1I8NTC6 A0A0T6BDA4 A0A1I8MIX7 A0A034WBC1 W8AVH1 A0A0K8UQH0 A0A0K8WCL8 A0A034WGI3 K7IWY2 A0A232FH95 A0A146KS09 A0A0A9W5I5 A0A023F522 A0A1Q3F413 T1IC53 A0A1A9XMX5 U5EVF7 A0A1W4VQ12 A0A182R054 Q94545 A0A182H8T1 A0A182HC27 B4PVR5 A0A182K860 A0A182F8E5 A0A182YEV3 B4HH49 A0A182UW56 A0A182TYK0 A0A182WVF3 Q5TT37 A0A182I932 A0A182W2Z6 W5JKQ7 A0A182KXX9 A0A182PU00 B3P708 B4GPG0 B5DXS8 A0A2M4BK60 A0A182NCL0 A0A182R8D4 A0A3B0K8F0 Q0IFZ7 B4K509 B4LW29 A0A1B0A8U9 A0A1A9UQ50 I3RSF7 B4N8U0 B4JEJ8 A0A0L7QYX7 B3M048 B0WEX2 A0A0P6G913 A0A0P6ADR8 A0A1B0ET16 A0A336M6A2 A0A084WR33 A0A0P6JWD4 A0A1S3D0C8 A0A0M5JC78 A0A182MP34 A0A182J148 A0A069DVC9 A0A0P6H0J6 A0A2H8THW3 A0A0P6B7C5 E9G3M1 A0A1D2N0E0 E0W163 A0A226EVX8 A0A336LH73
E9J271 A0A151I3D9 A0A151IW66 E2ABY2 A0A026X3K3 A0A151WIR0 F4WMV2 A0A195FV15 A0A2J7QW12 A0A0N0BGY4 A0A067R8L5 A0A1B6ERW4 A0A0C9RK78 A0A0C9RK43 A0A088A739 A0A2A3E6E0 A0A0L0C6Q0 A0A1B6LI47 A0A154PC03 A0A347ZJ80 E2BJ62 A0A1I8NTC6 A0A0T6BDA4 A0A1I8MIX7 A0A034WBC1 W8AVH1 A0A0K8UQH0 A0A0K8WCL8 A0A034WGI3 K7IWY2 A0A232FH95 A0A146KS09 A0A0A9W5I5 A0A023F522 A0A1Q3F413 T1IC53 A0A1A9XMX5 U5EVF7 A0A1W4VQ12 A0A182R054 Q94545 A0A182H8T1 A0A182HC27 B4PVR5 A0A182K860 A0A182F8E5 A0A182YEV3 B4HH49 A0A182UW56 A0A182TYK0 A0A182WVF3 Q5TT37 A0A182I932 A0A182W2Z6 W5JKQ7 A0A182KXX9 A0A182PU00 B3P708 B4GPG0 B5DXS8 A0A2M4BK60 A0A182NCL0 A0A182R8D4 A0A3B0K8F0 Q0IFZ7 B4K509 B4LW29 A0A1B0A8U9 A0A1A9UQ50 I3RSF7 B4N8U0 B4JEJ8 A0A0L7QYX7 B3M048 B0WEX2 A0A0P6G913 A0A0P6ADR8 A0A1B0ET16 A0A336M6A2 A0A084WR33 A0A0P6JWD4 A0A1S3D0C8 A0A0M5JC78 A0A182MP34 A0A182J148 A0A069DVC9 A0A0P6H0J6 A0A2H8THW3 A0A0P6B7C5 E9G3M1 A0A1D2N0E0 E0W163 A0A226EVX8 A0A336LH73
Pubmed
19121390
26354079
22118469
28004739
18362917
19820115
+ More
21282665 20798317 24508170 30249741 21719571 24845553 26108605 25315136 25348373 24495485 20075255 28648823 26823975 25401762 25474469 8889525 10731132 12537572 12537569 8555105 12068954 12626737 15371308 15280236 17466076 21694722 21875999 21310711 23197473 26483478 17994087 17550304 25244985 12364791 14747013 17210077 20920257 23761445 20966253 15632085 17510324 22328714 24438588 26334808 21292972 27289101 20566863
21282665 20798317 24508170 30249741 21719571 24845553 26108605 25315136 25348373 24495485 20075255 28648823 26823975 25401762 25474469 8889525 10731132 12537572 12537569 8555105 12068954 12626737 15371308 15280236 17466076 21694722 21875999 21310711 23197473 26483478 17994087 17550304 25244985 12364791 14747013 17210077 20920257 23761445 20966253 15632085 17510324 22328714 24438588 26334808 21292972 27289101 20566863
EMBL
BABH01023354
BABH01023355
ODYU01007214
SOQ49798.1
KQ459185
KPJ03388.1
+ More
AGBW02014406 OWR41724.1 GEZM01028888 JAV86210.1 KQ971377 EEZ97384.2 GL767820 EFZ13081.1 KQ976482 KYM83725.1 KQ980882 KYN11937.1 GL438386 EFN69077.1 KK107019 QOIP01000007 EZA62643.1 RLU20024.1 KQ983089 KYQ47625.1 GL888223 EGI64422.1 KQ981261 KYN44132.1 NEVH01009768 PNF32764.1 KQ435762 KOX75454.1 KK852667 KDR18916.1 GECZ01029097 GECZ01012982 JAS40672.1 JAS56787.1 GBYB01007241 GBYB01008471 GBYB01008472 JAG77008.1 JAG78238.1 JAG78239.1 GBYB01008580 JAG78347.1 KZ288353 PBC27280.1 JRES01000835 KNC27911.1 GEBQ01016703 JAT23274.1 KQ434869 KZC09415.1 FX985648 BBA84386.1 GL448558 EFN84212.1 LJIG01001626 KRT85314.1 GAKP01006086 GAKP01006076 JAC52866.1 GAMC01017872 JAB88683.1 GDHF01023581 JAI28733.1 GDHF01003714 JAI48600.1 GAKP01006079 GAKP01006069 JAC52883.1 AAZX01004124 NNAY01000194 OXU30104.1 GDHC01019368 JAP99260.1 GBHO01039902 JAG03702.1 GBBI01002573 JAC16139.1 GFDL01012750 JAV22295.1 ACPB03004429 GANO01003425 JAB56446.1 AXCN02001184 U73809 AE014297 AY051803 BT011328 BT120264 AAC47328.2 AAK93227.1 AAO41602.1 AAR96120.1 JXUM01119205 KQ566265 KXJ70267.1 JXUM01126286 JXUM01126287 JXUM01126288 CM000160 EDW98914.2 CH480815 EDW43525.1 AAAB01008888 EAL40623.3 APCN01003167 ADMH02001252 ETN63480.1 CH954182 EDV53828.1 CH479186 EDW39043.1 CM000070 EDY67519.1 GGFJ01004200 MBW53341.1 OUUW01000005 SPP81301.1 CH477281 EAT44927.1 CH933806 EDW13980.2 CH940650 EDW66534.1 JQ611724 AFK29242.1 CH964232 EDW81541.1 CH916369 EDV93129.1 KQ414686 KOC63766.1 CH902617 EDV42005.1 DS231912 EDS25830.1 GDIQ01039557 JAN55180.1 GDIP01044059 LRGB01001574 JAM59656.1 KZS11620.1 AJWK01005374 AJWK01005375 UFQT01000615 SSX25786.1 ATLV01025911 KE525402 KFB52677.1 GDIQ01006018 JAN88719.1 CP012526 ALC45918.1 AXCM01000016 GBGD01001118 JAC87771.1 GDIQ01036575 JAN58162.1 GFXV01001487 MBW13292.1 GDIP01019377 JAM84338.1 GL732531 EFX85961.1 LJIJ01000336 ODM98671.1 DS235866 EEB19369.1 LNIX01000001 OXA61679.1 UFQS01004023 UFQT01004023 SSX16107.1 SSX35436.1
AGBW02014406 OWR41724.1 GEZM01028888 JAV86210.1 KQ971377 EEZ97384.2 GL767820 EFZ13081.1 KQ976482 KYM83725.1 KQ980882 KYN11937.1 GL438386 EFN69077.1 KK107019 QOIP01000007 EZA62643.1 RLU20024.1 KQ983089 KYQ47625.1 GL888223 EGI64422.1 KQ981261 KYN44132.1 NEVH01009768 PNF32764.1 KQ435762 KOX75454.1 KK852667 KDR18916.1 GECZ01029097 GECZ01012982 JAS40672.1 JAS56787.1 GBYB01007241 GBYB01008471 GBYB01008472 JAG77008.1 JAG78238.1 JAG78239.1 GBYB01008580 JAG78347.1 KZ288353 PBC27280.1 JRES01000835 KNC27911.1 GEBQ01016703 JAT23274.1 KQ434869 KZC09415.1 FX985648 BBA84386.1 GL448558 EFN84212.1 LJIG01001626 KRT85314.1 GAKP01006086 GAKP01006076 JAC52866.1 GAMC01017872 JAB88683.1 GDHF01023581 JAI28733.1 GDHF01003714 JAI48600.1 GAKP01006079 GAKP01006069 JAC52883.1 AAZX01004124 NNAY01000194 OXU30104.1 GDHC01019368 JAP99260.1 GBHO01039902 JAG03702.1 GBBI01002573 JAC16139.1 GFDL01012750 JAV22295.1 ACPB03004429 GANO01003425 JAB56446.1 AXCN02001184 U73809 AE014297 AY051803 BT011328 BT120264 AAC47328.2 AAK93227.1 AAO41602.1 AAR96120.1 JXUM01119205 KQ566265 KXJ70267.1 JXUM01126286 JXUM01126287 JXUM01126288 CM000160 EDW98914.2 CH480815 EDW43525.1 AAAB01008888 EAL40623.3 APCN01003167 ADMH02001252 ETN63480.1 CH954182 EDV53828.1 CH479186 EDW39043.1 CM000070 EDY67519.1 GGFJ01004200 MBW53341.1 OUUW01000005 SPP81301.1 CH477281 EAT44927.1 CH933806 EDW13980.2 CH940650 EDW66534.1 JQ611724 AFK29242.1 CH964232 EDW81541.1 CH916369 EDV93129.1 KQ414686 KOC63766.1 CH902617 EDV42005.1 DS231912 EDS25830.1 GDIQ01039557 JAN55180.1 GDIP01044059 LRGB01001574 JAM59656.1 KZS11620.1 AJWK01005374 AJWK01005375 UFQT01000615 SSX25786.1 ATLV01025911 KE525402 KFB52677.1 GDIQ01006018 JAN88719.1 CP012526 ALC45918.1 AXCM01000016 GBGD01001118 JAC87771.1 GDIQ01036575 JAN58162.1 GFXV01001487 MBW13292.1 GDIP01019377 JAM84338.1 GL732531 EFX85961.1 LJIJ01000336 ODM98671.1 DS235866 EEB19369.1 LNIX01000001 OXA61679.1 UFQS01004023 UFQT01004023 SSX16107.1 SSX35436.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000007266
UP000078540
UP000078492
+ More
UP000000311 UP000053097 UP000279307 UP000075809 UP000007755 UP000078541 UP000235965 UP000053105 UP000027135 UP000005203 UP000242457 UP000037069 UP000076502 UP000008237 UP000095300 UP000095301 UP000002358 UP000215335 UP000015103 UP000092443 UP000192221 UP000075886 UP000000803 UP000069940 UP000249989 UP000002282 UP000075881 UP000069272 UP000076408 UP000001292 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000075920 UP000000673 UP000075882 UP000075885 UP000008711 UP000008744 UP000001819 UP000075884 UP000075900 UP000268350 UP000008820 UP000009192 UP000008792 UP000092445 UP000078200 UP000007798 UP000001070 UP000053825 UP000007801 UP000002320 UP000076858 UP000092461 UP000030765 UP000079169 UP000092553 UP000075883 UP000075880 UP000000305 UP000094527 UP000009046 UP000198287
UP000000311 UP000053097 UP000279307 UP000075809 UP000007755 UP000078541 UP000235965 UP000053105 UP000027135 UP000005203 UP000242457 UP000037069 UP000076502 UP000008237 UP000095300 UP000095301 UP000002358 UP000215335 UP000015103 UP000092443 UP000192221 UP000075886 UP000000803 UP000069940 UP000249989 UP000002282 UP000075881 UP000069272 UP000076408 UP000001292 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000075920 UP000000673 UP000075882 UP000075885 UP000008711 UP000008744 UP000001819 UP000075884 UP000075900 UP000268350 UP000008820 UP000009192 UP000008792 UP000092445 UP000078200 UP000007798 UP000001070 UP000053825 UP000007801 UP000002320 UP000076858 UP000092461 UP000030765 UP000079169 UP000092553 UP000075883 UP000075880 UP000000305 UP000094527 UP000009046 UP000198287
Pfam
PF00622 SPRY
Interpro
ProteinModelPortal
H9JES8
A0A2H1W9Q6
A0A194QCW9
A0A212EJS5
A0A1Y1MNB3
D6X3Q8
+ More
E9J271 A0A151I3D9 A0A151IW66 E2ABY2 A0A026X3K3 A0A151WIR0 F4WMV2 A0A195FV15 A0A2J7QW12 A0A0N0BGY4 A0A067R8L5 A0A1B6ERW4 A0A0C9RK78 A0A0C9RK43 A0A088A739 A0A2A3E6E0 A0A0L0C6Q0 A0A1B6LI47 A0A154PC03 A0A347ZJ80 E2BJ62 A0A1I8NTC6 A0A0T6BDA4 A0A1I8MIX7 A0A034WBC1 W8AVH1 A0A0K8UQH0 A0A0K8WCL8 A0A034WGI3 K7IWY2 A0A232FH95 A0A146KS09 A0A0A9W5I5 A0A023F522 A0A1Q3F413 T1IC53 A0A1A9XMX5 U5EVF7 A0A1W4VQ12 A0A182R054 Q94545 A0A182H8T1 A0A182HC27 B4PVR5 A0A182K860 A0A182F8E5 A0A182YEV3 B4HH49 A0A182UW56 A0A182TYK0 A0A182WVF3 Q5TT37 A0A182I932 A0A182W2Z6 W5JKQ7 A0A182KXX9 A0A182PU00 B3P708 B4GPG0 B5DXS8 A0A2M4BK60 A0A182NCL0 A0A182R8D4 A0A3B0K8F0 Q0IFZ7 B4K509 B4LW29 A0A1B0A8U9 A0A1A9UQ50 I3RSF7 B4N8U0 B4JEJ8 A0A0L7QYX7 B3M048 B0WEX2 A0A0P6G913 A0A0P6ADR8 A0A1B0ET16 A0A336M6A2 A0A084WR33 A0A0P6JWD4 A0A1S3D0C8 A0A0M5JC78 A0A182MP34 A0A182J148 A0A069DVC9 A0A0P6H0J6 A0A2H8THW3 A0A0P6B7C5 E9G3M1 A0A1D2N0E0 E0W163 A0A226EVX8 A0A336LH73
E9J271 A0A151I3D9 A0A151IW66 E2ABY2 A0A026X3K3 A0A151WIR0 F4WMV2 A0A195FV15 A0A2J7QW12 A0A0N0BGY4 A0A067R8L5 A0A1B6ERW4 A0A0C9RK78 A0A0C9RK43 A0A088A739 A0A2A3E6E0 A0A0L0C6Q0 A0A1B6LI47 A0A154PC03 A0A347ZJ80 E2BJ62 A0A1I8NTC6 A0A0T6BDA4 A0A1I8MIX7 A0A034WBC1 W8AVH1 A0A0K8UQH0 A0A0K8WCL8 A0A034WGI3 K7IWY2 A0A232FH95 A0A146KS09 A0A0A9W5I5 A0A023F522 A0A1Q3F413 T1IC53 A0A1A9XMX5 U5EVF7 A0A1W4VQ12 A0A182R054 Q94545 A0A182H8T1 A0A182HC27 B4PVR5 A0A182K860 A0A182F8E5 A0A182YEV3 B4HH49 A0A182UW56 A0A182TYK0 A0A182WVF3 Q5TT37 A0A182I932 A0A182W2Z6 W5JKQ7 A0A182KXX9 A0A182PU00 B3P708 B4GPG0 B5DXS8 A0A2M4BK60 A0A182NCL0 A0A182R8D4 A0A3B0K8F0 Q0IFZ7 B4K509 B4LW29 A0A1B0A8U9 A0A1A9UQ50 I3RSF7 B4N8U0 B4JEJ8 A0A0L7QYX7 B3M048 B0WEX2 A0A0P6G913 A0A0P6ADR8 A0A1B0ET16 A0A336M6A2 A0A084WR33 A0A0P6JWD4 A0A1S3D0C8 A0A0M5JC78 A0A182MP34 A0A182J148 A0A069DVC9 A0A0P6H0J6 A0A2H8THW3 A0A0P6B7C5 E9G3M1 A0A1D2N0E0 E0W163 A0A226EVX8 A0A336LH73
PDB
6E2H
E-value=3.07125e-59,
Score=580
Ontologies
GO
GO:0048188
GO:0051568
GO:0046872
GO:0008168
GO:0044212
GO:0006351
GO:0005634
GO:0044666
GO:0007420
GO:0007444
GO:0051571
GO:0044665
GO:0003677
GO:0007474
GO:0010628
GO:0035209
GO:0048813
GO:0006357
GO:0048096
GO:0033128
GO:0005703
GO:0019899
GO:0035075
GO:0005700
GO:0002168
GO:0007476
GO:0010629
GO:0005515
GO:0008270
GO:0016021
GO:0005524
GO:0005886
GO:0007166
GO:0006032
GO:0016742
GO:0003824
PANTHER
Topology
Subcellular location
Nucleus
Accumulates on salivary gland polytene chromosomes. With evidence from 8 publications.
Chromosome Accumulates on salivary gland polytene chromosomes. With evidence from 8 publications.
Chromosome Accumulates on salivary gland polytene chromosomes. With evidence from 8 publications.
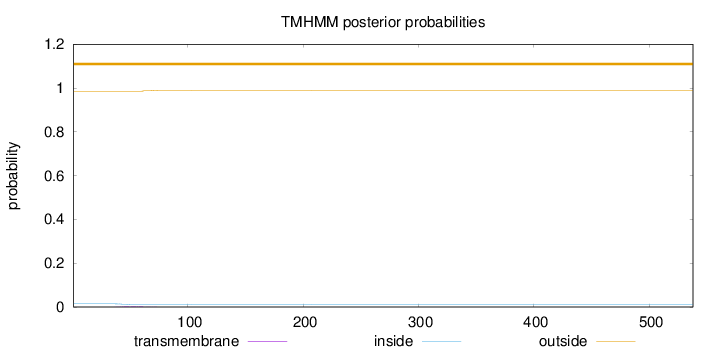
Length:
538
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0730700000000002
Exp number, first 60 AAs:
0.06051
Total prob of N-in:
0.01485
outside
1 - 538
Population Genetic Test Statistics
Pi
326.042987
Theta
183.667858
Tajima's D
-1.163414
CLR
1133.186524
CSRT
0.109094545272736
Interpretation
Uncertain
