Gene
KWMTBOMO04866 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008047
Annotation
PREDICTED:_bolA-like_protein_DDB_G0274169_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.216 Mitochondrial Reliability : 1.31
Sequence
CDS
ATGTCATCCGCATCAGGAATAGTCGAAAGTACAATAAGAAATAAATTATTAACAATTTTAGAATCAAAGCATGTCCAGGTGATAAATGAATCCTACATGCACAATGTACCAAAAGGAGCTGAAACACACTTCAAAGTAGTAGTTGTTTCAGAAAAATTTGATGGTTTACCTCTTATAAAAAGACATCGTTTAGTCAATGAACTATTGGCAGAAGAACTTCAAAATGGTGTACATGCACTATCAATAGTAGCAAAAACACCTAAACAATGGGAGGAAAGTGATAAACTGGTTGAGAACAGTCCCAACTGTAGAGGAGGATTTGGAATGGCTTATTTGGAGCCGACGAGCGACAATCCGCTGGCTGAGCTCTTCTTGCAATAG
Protein
MSSASGIVESTIRNKLLTILESKHVQVINESYMHNVPKGAETHFKVVVVSEKFDGLPLIKRHRLVNELLAEELQNGVHALSIVAKTPKQWEESDKLVENSPNCRGGFGMAYLEPTSDNPLAELFLQ
Summary
Similarity
Belongs to the BolA/IbaG family.
Belongs to the AB hydrolase superfamily. Lipase family.
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
A0A2W1BTJ9
S4NRV8
A0A194QKD3
A0A212EJQ0
A0A2H1W9S3
A0A1W4WH41
+ More
A0A194QD92 A0A1W4WSX7 A0A0L7KLR5 A0A232FHR9 K7IWY3 A0A139WB15 A0A2P8XV11 A0A151WIH8 A0A0P4W3F9 A0A026X321 A0A0V0G8V9 A0A069DNR9 A0A3L8DHU6 E9J270 J9JM94 A0A023F0C1 E2BJ63 A0A0P4VQ36 A0A195FUN2 T1I5Z6 A0A158NXX5 A0A1Y1KF03 A0A2J7RQE3 E9FYS5 A0A067QQZ7 A0A1Y1K9Q3 A0A0P5VGJ8 A0A3M6VVI5 A0A0P5IEW2 A0A0A9VZ93 G4ZLH3 W2IGS6 W2KQR4 A0A0W8CZV0 W2WHU3 V9EP38 W2PTW7 A0A080ZPK3 V4BKI0 A0A0L7QYS2 E2ABY1 A0A182K859 A0A182NCL1 A0A182UR52 A0A182PTZ9 A0A1Q3FB07 Q0IFZ6 Q7QAG7 J3JW03 K3X665 A0A329SYD8 A0A182R8D5 A0A023EES1 A0A182H8T0 B0WEX1 A0A182WVF2 A0A023EGI1 A0A023EE26 A0A2H8TZE4 A0A2D4BZF7 A0A3F2YVG0 A0A182I931 C3Z5H6 A0A0P6JWJ7 A0A182UAB4 A0A225UUH1 A0A182W2Z7 A0A1S6GL27 M4BAW4 D0NU59 A0A084WR32 A0A182YEV4 A0A182ISP9 Q6VFI5 Q6VFI3 A0A2A3E7W8 A0A182Q6T3 H3AJZ9 A0A1I8PLQ4 U5ESX8 A0A0P1A6A6 E0W122 W8BVG2 A0A2M4CYQ4 A0A1S3JWZ4 A0A336KKG8 A0A024UJH8 A0A336N3H9 A0A2M4AW52 A0A2M4AW43 B3RW48 W5JHD7
A0A194QD92 A0A1W4WSX7 A0A0L7KLR5 A0A232FHR9 K7IWY3 A0A139WB15 A0A2P8XV11 A0A151WIH8 A0A0P4W3F9 A0A026X321 A0A0V0G8V9 A0A069DNR9 A0A3L8DHU6 E9J270 J9JM94 A0A023F0C1 E2BJ63 A0A0P4VQ36 A0A195FUN2 T1I5Z6 A0A158NXX5 A0A1Y1KF03 A0A2J7RQE3 E9FYS5 A0A067QQZ7 A0A1Y1K9Q3 A0A0P5VGJ8 A0A3M6VVI5 A0A0P5IEW2 A0A0A9VZ93 G4ZLH3 W2IGS6 W2KQR4 A0A0W8CZV0 W2WHU3 V9EP38 W2PTW7 A0A080ZPK3 V4BKI0 A0A0L7QYS2 E2ABY1 A0A182K859 A0A182NCL1 A0A182UR52 A0A182PTZ9 A0A1Q3FB07 Q0IFZ6 Q7QAG7 J3JW03 K3X665 A0A329SYD8 A0A182R8D5 A0A023EES1 A0A182H8T0 B0WEX1 A0A182WVF2 A0A023EGI1 A0A023EE26 A0A2H8TZE4 A0A2D4BZF7 A0A3F2YVG0 A0A182I931 C3Z5H6 A0A0P6JWJ7 A0A182UAB4 A0A225UUH1 A0A182W2Z7 A0A1S6GL27 M4BAW4 D0NU59 A0A084WR32 A0A182YEV4 A0A182ISP9 Q6VFI5 Q6VFI3 A0A2A3E7W8 A0A182Q6T3 H3AJZ9 A0A1I8PLQ4 U5ESX8 A0A0P1A6A6 E0W122 W8BVG2 A0A2M4CYQ4 A0A1S3JWZ4 A0A336KKG8 A0A024UJH8 A0A336N3H9 A0A2M4AW52 A0A2M4AW43 B3RW48 W5JHD7
Pubmed
28756777
23622113
26354079
22118469
26227816
28648823
+ More
20075255 18362917 19820115 29403074 24508170 26334808 30249741 21282665 25474469 20798317 27129103 21347285 28004739 21292972 24845553 25401762 26823975 16946064 23254933 17510324 12364791 14747013 17210077 22516182 23537049 24945155 26483478 18563158 28043022 21148394 19741609 24438588 25244985 9215903 20566863 24495485 18719581 20920257 23761445
20075255 18362917 19820115 29403074 24508170 26334808 30249741 21282665 25474469 20798317 27129103 21347285 28004739 21292972 24845553 25401762 26823975 16946064 23254933 17510324 12364791 14747013 17210077 22516182 23537049 24945155 26483478 18563158 28043022 21148394 19741609 24438588 25244985 9215903 20566863 24495485 18719581 20920257 23761445
EMBL
KZ149969
PZC76100.1
GAIX01011034
JAA81526.1
KQ461198
KPJ05992.1
+ More
AGBW02014406 OWR41723.1 ODYU01007214 SOQ49797.1 KQ459185 KPJ03389.1 JTDY01009250 KOB63844.1 NNAY01000194 OXU30103.1 AAZX01004124 KQ971377 KYB25102.1 PYGN01001304 PSN35842.1 KQ983089 KYQ47626.1 GDRN01075576 JAI63038.1 KK107019 EZA62642.1 GECL01001514 JAP04610.1 GBGD01003583 JAC85306.1 QOIP01000007 RLU20025.1 GL767820 EFZ13075.1 ABLF02034197 GBBI01004423 JAC14289.1 GL448558 EFN84213.1 GDKW01003305 JAI53290.1 KQ981261 KYN44131.1 ACPB03008320 ADTU01003678 GEZM01088437 GEZM01088434 GEZM01088431 GEZM01088430 JAV58205.1 NEVH01001340 PNF43051.1 GL732527 EFX87587.1 KK853125 KDR11022.1 GEZM01088436 GEZM01088435 GEZM01088433 GEZM01088432 JAV58199.1 GDIP01100217 LRGB01000930 JAM03498.1 KZS14983.1 QLLG01000001 QKXF01000113 RMX70492.1 RQM16605.1 GDIQ01214376 JAK37349.1 GBHO01045669 GBRD01001886 GBRD01001883 GDHC01016071 JAF97934.1 JAG63935.1 JAQ02558.1 JH159155 EGZ16255.1 KI687779 KI674535 ETK79943.1 ETL33361.1 KI681208 KI694461 ETL86670.1 ETM39842.1 LNFO01003045 LNFP01000778 KUF83907.1 KUF89626.1 ANIX01002838 ETP09683.1 ANIZ01002513 ETI39837.1 KI669604 ETN04367.1 ANJA01002629 ETO68564.1 KB202619 ESO89079.1 KQ414686 KOC63765.1 GL438386 EFN69076.1 GFDL01010305 JAV24740.1 CH477281 EAT44928.1 AAAB01008888 EAA08757.5 APGK01057982 APGK01057983 BT127421 KB741285 AEE62383.1 ENN70740.1 GL376588 MJFZ01000039 RAW40956.1 GAPW01005721 JAC07877.1 JXUM01119205 KQ566265 KXJ70266.1 DS231912 EDS25829.1 GAPW01005759 JAC07839.1 GAPW01005740 JAC07858.1 GFXV01007087 MBW18892.1 BBXB01000093 BBXB01000534 GAX98749.1 GAY06037.1 AXCM01000016 APCN01003167 GG666583 EEN52089.1 EEN52110.1 GDIQ01005744 JAN88993.1 NBNE01011232 OWY96780.1 KY314132 AQS22566.1 JH598083 DS028162 EEY65183.1 ATLV01025911 KE525402 KFB52676.1 AY341151 AAR13715.1 AY341152 AY341153 AY341154 AY341155 AY341156 AAR13717.1 AAR13718.1 KZ288353 PBC27279.1 AXCN02001184 AFYH01035194 GANO01002173 JAB57698.1 CCYD01000109 CEG36113.1 DS235864 EEB19328.1 GAMC01009254 JAB97301.1 GGFL01006249 MBW70427.1 UFQS01000615 UFQT01000615 SSX05424.1 SSX25784.1 KI913956 ETW06007.1 UFQT01004023 SSX35437.1 GGFK01011693 MBW45014.1 GGFK01011692 MBW45013.1 DS985244 EDV26115.1 ADMH02001252 ETN63481.1
AGBW02014406 OWR41723.1 ODYU01007214 SOQ49797.1 KQ459185 KPJ03389.1 JTDY01009250 KOB63844.1 NNAY01000194 OXU30103.1 AAZX01004124 KQ971377 KYB25102.1 PYGN01001304 PSN35842.1 KQ983089 KYQ47626.1 GDRN01075576 JAI63038.1 KK107019 EZA62642.1 GECL01001514 JAP04610.1 GBGD01003583 JAC85306.1 QOIP01000007 RLU20025.1 GL767820 EFZ13075.1 ABLF02034197 GBBI01004423 JAC14289.1 GL448558 EFN84213.1 GDKW01003305 JAI53290.1 KQ981261 KYN44131.1 ACPB03008320 ADTU01003678 GEZM01088437 GEZM01088434 GEZM01088431 GEZM01088430 JAV58205.1 NEVH01001340 PNF43051.1 GL732527 EFX87587.1 KK853125 KDR11022.1 GEZM01088436 GEZM01088435 GEZM01088433 GEZM01088432 JAV58199.1 GDIP01100217 LRGB01000930 JAM03498.1 KZS14983.1 QLLG01000001 QKXF01000113 RMX70492.1 RQM16605.1 GDIQ01214376 JAK37349.1 GBHO01045669 GBRD01001886 GBRD01001883 GDHC01016071 JAF97934.1 JAG63935.1 JAQ02558.1 JH159155 EGZ16255.1 KI687779 KI674535 ETK79943.1 ETL33361.1 KI681208 KI694461 ETL86670.1 ETM39842.1 LNFO01003045 LNFP01000778 KUF83907.1 KUF89626.1 ANIX01002838 ETP09683.1 ANIZ01002513 ETI39837.1 KI669604 ETN04367.1 ANJA01002629 ETO68564.1 KB202619 ESO89079.1 KQ414686 KOC63765.1 GL438386 EFN69076.1 GFDL01010305 JAV24740.1 CH477281 EAT44928.1 AAAB01008888 EAA08757.5 APGK01057982 APGK01057983 BT127421 KB741285 AEE62383.1 ENN70740.1 GL376588 MJFZ01000039 RAW40956.1 GAPW01005721 JAC07877.1 JXUM01119205 KQ566265 KXJ70266.1 DS231912 EDS25829.1 GAPW01005759 JAC07839.1 GAPW01005740 JAC07858.1 GFXV01007087 MBW18892.1 BBXB01000093 BBXB01000534 GAX98749.1 GAY06037.1 AXCM01000016 APCN01003167 GG666583 EEN52089.1 EEN52110.1 GDIQ01005744 JAN88993.1 NBNE01011232 OWY96780.1 KY314132 AQS22566.1 JH598083 DS028162 EEY65183.1 ATLV01025911 KE525402 KFB52676.1 AY341151 AAR13715.1 AY341152 AY341153 AY341154 AY341155 AY341156 AAR13717.1 AAR13718.1 KZ288353 PBC27279.1 AXCN02001184 AFYH01035194 GANO01002173 JAB57698.1 CCYD01000109 CEG36113.1 DS235864 EEB19328.1 GAMC01009254 JAB97301.1 GGFL01006249 MBW70427.1 UFQS01000615 UFQT01000615 SSX05424.1 SSX25784.1 KI913956 ETW06007.1 UFQT01004023 SSX35437.1 GGFK01011693 MBW45014.1 GGFK01011692 MBW45013.1 DS985244 EDV26115.1 ADMH02001252 ETN63481.1
Proteomes
UP000053240
UP000007151
UP000192223
UP000053268
UP000037510
UP000215335
+ More
UP000002358 UP000007266 UP000245037 UP000075809 UP000053097 UP000279307 UP000007819 UP000008237 UP000078541 UP000015103 UP000005205 UP000235965 UP000000305 UP000027135 UP000076858 UP000282087 UP000286097 UP000002640 UP000053236 UP000053864 UP000054423 UP000054532 UP000052943 UP000018958 UP000018721 UP000018817 UP000028582 UP000030746 UP000053825 UP000000311 UP000075881 UP000075884 UP000075903 UP000075885 UP000008820 UP000007062 UP000019118 UP000251314 UP000075900 UP000069940 UP000249989 UP000002320 UP000076407 UP000054177 UP000075883 UP000075840 UP000001554 UP000075902 UP000198211 UP000075920 UP000011713 UP000006643 UP000030765 UP000076408 UP000075880 UP000242457 UP000075886 UP000008672 UP000095300 UP000054928 UP000009046 UP000085678 UP000024375 UP000009022 UP000000673
UP000002358 UP000007266 UP000245037 UP000075809 UP000053097 UP000279307 UP000007819 UP000008237 UP000078541 UP000015103 UP000005205 UP000235965 UP000000305 UP000027135 UP000076858 UP000282087 UP000286097 UP000002640 UP000053236 UP000053864 UP000054423 UP000054532 UP000052943 UP000018958 UP000018721 UP000018817 UP000028582 UP000030746 UP000053825 UP000000311 UP000075881 UP000075884 UP000075903 UP000075885 UP000008820 UP000007062 UP000019118 UP000251314 UP000075900 UP000069940 UP000249989 UP000002320 UP000076407 UP000054177 UP000075883 UP000075840 UP000001554 UP000075902 UP000198211 UP000075920 UP000011713 UP000006643 UP000030765 UP000076408 UP000075880 UP000242457 UP000075886 UP000008672 UP000095300 UP000054928 UP000009046 UP000085678 UP000024375 UP000009022 UP000000673
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BTJ9
S4NRV8
A0A194QKD3
A0A212EJQ0
A0A2H1W9S3
A0A1W4WH41
+ More
A0A194QD92 A0A1W4WSX7 A0A0L7KLR5 A0A232FHR9 K7IWY3 A0A139WB15 A0A2P8XV11 A0A151WIH8 A0A0P4W3F9 A0A026X321 A0A0V0G8V9 A0A069DNR9 A0A3L8DHU6 E9J270 J9JM94 A0A023F0C1 E2BJ63 A0A0P4VQ36 A0A195FUN2 T1I5Z6 A0A158NXX5 A0A1Y1KF03 A0A2J7RQE3 E9FYS5 A0A067QQZ7 A0A1Y1K9Q3 A0A0P5VGJ8 A0A3M6VVI5 A0A0P5IEW2 A0A0A9VZ93 G4ZLH3 W2IGS6 W2KQR4 A0A0W8CZV0 W2WHU3 V9EP38 W2PTW7 A0A080ZPK3 V4BKI0 A0A0L7QYS2 E2ABY1 A0A182K859 A0A182NCL1 A0A182UR52 A0A182PTZ9 A0A1Q3FB07 Q0IFZ6 Q7QAG7 J3JW03 K3X665 A0A329SYD8 A0A182R8D5 A0A023EES1 A0A182H8T0 B0WEX1 A0A182WVF2 A0A023EGI1 A0A023EE26 A0A2H8TZE4 A0A2D4BZF7 A0A3F2YVG0 A0A182I931 C3Z5H6 A0A0P6JWJ7 A0A182UAB4 A0A225UUH1 A0A182W2Z7 A0A1S6GL27 M4BAW4 D0NU59 A0A084WR32 A0A182YEV4 A0A182ISP9 Q6VFI5 Q6VFI3 A0A2A3E7W8 A0A182Q6T3 H3AJZ9 A0A1I8PLQ4 U5ESX8 A0A0P1A6A6 E0W122 W8BVG2 A0A2M4CYQ4 A0A1S3JWZ4 A0A336KKG8 A0A024UJH8 A0A336N3H9 A0A2M4AW52 A0A2M4AW43 B3RW48 W5JHD7
A0A194QD92 A0A1W4WSX7 A0A0L7KLR5 A0A232FHR9 K7IWY3 A0A139WB15 A0A2P8XV11 A0A151WIH8 A0A0P4W3F9 A0A026X321 A0A0V0G8V9 A0A069DNR9 A0A3L8DHU6 E9J270 J9JM94 A0A023F0C1 E2BJ63 A0A0P4VQ36 A0A195FUN2 T1I5Z6 A0A158NXX5 A0A1Y1KF03 A0A2J7RQE3 E9FYS5 A0A067QQZ7 A0A1Y1K9Q3 A0A0P5VGJ8 A0A3M6VVI5 A0A0P5IEW2 A0A0A9VZ93 G4ZLH3 W2IGS6 W2KQR4 A0A0W8CZV0 W2WHU3 V9EP38 W2PTW7 A0A080ZPK3 V4BKI0 A0A0L7QYS2 E2ABY1 A0A182K859 A0A182NCL1 A0A182UR52 A0A182PTZ9 A0A1Q3FB07 Q0IFZ6 Q7QAG7 J3JW03 K3X665 A0A329SYD8 A0A182R8D5 A0A023EES1 A0A182H8T0 B0WEX1 A0A182WVF2 A0A023EGI1 A0A023EE26 A0A2H8TZE4 A0A2D4BZF7 A0A3F2YVG0 A0A182I931 C3Z5H6 A0A0P6JWJ7 A0A182UAB4 A0A225UUH1 A0A182W2Z7 A0A1S6GL27 M4BAW4 D0NU59 A0A084WR32 A0A182YEV4 A0A182ISP9 Q6VFI5 Q6VFI3 A0A2A3E7W8 A0A182Q6T3 H3AJZ9 A0A1I8PLQ4 U5ESX8 A0A0P1A6A6 E0W122 W8BVG2 A0A2M4CYQ4 A0A1S3JWZ4 A0A336KKG8 A0A024UJH8 A0A336N3H9 A0A2M4AW52 A0A2M4AW43 B3RW48 W5JHD7
PDB
5LCI
E-value=4.89371e-22,
Score=251
Ontologies
PANTHER
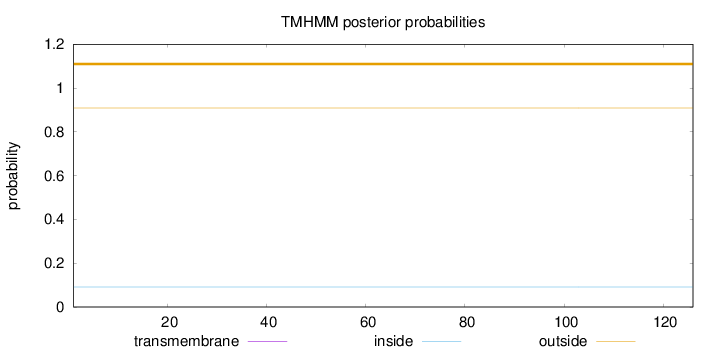
Topology
Subcellular location
Secreted
Length:
126
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09130
outside
1 - 126
Population Genetic Test Statistics
Pi
254.749926
Theta
218.927945
Tajima's D
0.728582
CLR
0.301407
CSRT
0.577271136443178
Interpretation
Uncertain
