Gene
KWMTBOMO04865
Pre Gene Modal
BGIBMGA008027
Annotation
PREDICTED:_sodium-_and_chloride-dependent_glycine_transporter_1-like_isoform_X2_[Amyelois_transitella]
Full name
Transporter
Location in the cell
PlasmaMembrane Reliability : 4.915
Sequence
CDS
ATGGCTAGTGGAGATTACACTATGAATAAGGGAGACGCAAATGGCAGGGCAACACAAAAAACAACCCAAGATGTGACGCCGGACAGGGCGAAGTGGGGCCGACCTCTGGAGTTCATTCTCGCTTGCCTGGGCTACGCCGTTGGCCTCGGAAACGTTTGGAGATTCCCATACTTGTCATATAAAAACGGCGGAGGTGCTTTCCTTCTTCCATTTTTCTTGATGTTGATTTTCATCGGTCTGCCAATCTTCTACTTGGAGCTGTACATAGGGCAGTACACGGGACTGGGTCCTCTAAAGGCATTCAGTGCGATCTCTCCATTCTTCAGTGGGCTCGGCTACTGCACTTTGGTCGTAATCAGCATCATTTCAATTTACTATATGATCATTGTGGCCTGGACGTTGTTCTACACGATTGTTTCTATTGGAGGAAACTTGGGCTGGGGATCTTGTGATAATGACTACAATACTGATTTTTGTTACAGCGGAGAATACGATAGGGTATGTCGTGAGAACAACACAGAGAACTACACGGACCTGACATTTTTTATGAGACAGTGCATGAGCATCGGTGAAATTTGTGAGAGGTCTGGGTTTGAGCCTCTTGATGACAAATATTGTTTAGATGGAAACGATCAAGTGCCTTGGTATTCTAATATCACAAGAGTATTGGCTTCAGAGGAATATTATAACGAGCGTGTATTGGGTCGCGGCGATGCGTCCTGGACTAACTGGGGCACTATACAGTGGCACCTCGTGGGGTGCTTATTTCTCTCTTGGGTCATCGCATTTTTGTGCGTCATAAAAGGTGTCCAGTCAGCAGGAAAAGTTGTGTACTTTACCGCACTGTTCCCCTACGTGATGTTGACGGCTCTGCTCATTCGGGGAGTGACTTTAGAAGGAGCTGTGGACGGAGTTCTATTTTACTTGACGCCGACTTGGTCAACCTTGCTGGAAGCCAAGGTCTGGGGTGACGCCGCATCACAAATATTCTACTCTTTTGGTGTGGCCTGCGGCTCCCTTGTCACTCTGGCTTCTTACAACAACTTCTATAACAACTGCCACTTCGATGCCGTCTTCGTCGCATTTGCCAACTTCTTGACTTCCATTTATGCTGGTTTTGCCATCTTCTCCGTTCTTGGATTTCAAGCTCAGCTCATGGGAGTCAGTGTTGATGACGTAGCAGAACAGGGTCCTGGCCTGGCATTCGTGGCATATCCAGAAGCTATACTGCAGATGCCAGTACCTCAGCTGTGGTCCATATTGTTCTTCTTCATGCTGTTCATTTTGGGTCTTGGCAGTCAGTTCGCTGGTATTGAAGCGATCAACACTGCTATAGTAGACCAGTGGCCGCATCTTAGGAAGAACTACTGGAGGGTAACAGCATTTACGTGTATGATGTGCTTCATATTGGGGATCCCGATGTGCTTCAGCGGAGGCGTCTACTTGTTGACTCTCCTCGATTGGAACACGGCCTCTTGGGCTATCCTTCTCATCGGTATGGCCGAGTGTGTGGCAGTATCTTGGAGTTATGGCATCAATCGCGCAATTAAAGATTTAGCCACGATGGGCATGACCCTAAATAAGTTTCTTCGATTCTACTGGAGGGCCGTTTGGACTGTCATCGTACCATTTGGTAGTATTGGTATTCTAGCTTTCATATTTTCGGACTGGACAGCTCCAGCTTACGAAGACTATGTTTTCCCGTTATTCGCTGATTTGCTGGGCTGGGCGGTTGGGCTGTCCACACTGGTATTATTCCCTGTAGGAGTTGCCTGGGCTATCTACCATGGATACAGAGGTAAGCAGTTGTTTACTCCAACTCCAGAGTGGGGACCGGCTGTGAAAGCACCTCCAGTAGAATCAACGCAGCTTGGAGGGCCTCACGACAATGGGGGCTACGTAATGTGA
Protein
MASGDYTMNKGDANGRATQKTTQDVTPDRAKWGRPLEFILACLGYAVGLGNVWRFPYLSYKNGGGAFLLPFFLMLIFIGLPIFYLELYIGQYTGLGPLKAFSAISPFFSGLGYCTLVVISIISIYYMIIVAWTLFYTIVSIGGNLGWGSCDNDYNTDFCYSGEYDRVCRENNTENYTDLTFFMRQCMSIGEICERSGFEPLDDKYCLDGNDQVPWYSNITRVLASEEYYNERVLGRGDASWTNWGTIQWHLVGCLFLSWVIAFLCVIKGVQSAGKVVYFTALFPYVMLTALLIRGVTLEGAVDGVLFYLTPTWSTLLEAKVWGDAASQIFYSFGVACGSLVTLASYNNFYNNCHFDAVFVAFANFLTSIYAGFAIFSVLGFQAQLMGVSVDDVAEQGPGLAFVAYPEAILQMPVPQLWSILFFFMLFILGLGSQFAGIEAINTAIVDQWPHLRKNYWRVTAFTCMMCFILGIPMCFSGGVYLLTLLDWNTASWAILLIGMAECVAVSWSYGINRAIKDLATMGMTLNKFLRFYWRAVWTVIVPFGSIGILAFIFSDWTAPAYEDYVFPLFADLLGWAVGLSTLVLFPVGVAWAIYHGYRGKQLFTPTPEWGPAVKAPPVESTQLGGPHDNGGYVM
Summary
Similarity
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Feature
chain Transporter
Uniprot
A0A2W1BM12
A0A2H1W6C2
A0A194QM12
A0A1E1WN46
A0A212FEF8
A0A3S2NLB0
+ More
A0A194QD56 A0A336LNJ2 A0A1J1IH02 A0A1L8DJN8 D6X3Q2 A0A139WB82 A0A088AFL2 A0A0C9Q1F8 E2AMG9 A0A1J1IQ76 A0A1J1I6T3 A0A1W4X5F8 A0A1J1I5D3 A0A3L8DPQ4 A0A232ELW2 A0A2J7PY70 A0A158NGE2 E2C2H1 U4TUP7 N6T994 A0A1W4WUR5 A0A0M9AAF7 A0A1J1I1C2 A0A195E733 A0A1J1I1A1 A0A151XC17 F4X6Y2 A0A195F1H6 A0A151IQB6 E9J630 A0A195B2Q5 B7S8D1 A0A2P8Z7V4 A0A1B0CZ55 T1FPK8 A0A2P2I1R7 A0A1S3HS51 A0A1S3HPA4 K7IZ11 A0A147BI19 A0A131XN80 A0A0N8CX87 A0A0P6E398 A0A224YZI4 A0A232EZ04 K7INE2 A0A0P5E6X7 A0A0P5D1I1 A0A224Z4S9 A0A1S3JWF3 A0A067RIW3 A0A0P5AGP6 A0A0P5NG54 A0A194PDE3 A0A293MSK5 A0A087UJK6 A0A194RHH9 A0A087SZR9 A0A310S8J2 A0A1E1X7M4 A0A0K2TDT5 A0A0N8B201 A0A0N8A4H0 A0A0P4Z373 A0A0P5T7C5 A0A0P5II29 A0A2R5LA64 A0A0N8CJA7 A0A023GE63 A0A0P4Z993 A0A088AI46 A0A154PE44 A0A1A9WIQ9 A0A0P4Z6R4 A0A087T9D3 Q293F9 H9IUJ7 A0A2P6L6H6 W8B744 E2C7A6 B4MYD7 T1J119 A0A0P6DBF2 A0A1A9Y0M4 A0A212EKK6 A0A0N8EJE9 A0A2T7PMP6 E0VAE8 A0A1I8J6K1 A0A1S3INW9
A0A194QD56 A0A336LNJ2 A0A1J1IH02 A0A1L8DJN8 D6X3Q2 A0A139WB82 A0A088AFL2 A0A0C9Q1F8 E2AMG9 A0A1J1IQ76 A0A1J1I6T3 A0A1W4X5F8 A0A1J1I5D3 A0A3L8DPQ4 A0A232ELW2 A0A2J7PY70 A0A158NGE2 E2C2H1 U4TUP7 N6T994 A0A1W4WUR5 A0A0M9AAF7 A0A1J1I1C2 A0A195E733 A0A1J1I1A1 A0A151XC17 F4X6Y2 A0A195F1H6 A0A151IQB6 E9J630 A0A195B2Q5 B7S8D1 A0A2P8Z7V4 A0A1B0CZ55 T1FPK8 A0A2P2I1R7 A0A1S3HS51 A0A1S3HPA4 K7IZ11 A0A147BI19 A0A131XN80 A0A0N8CX87 A0A0P6E398 A0A224YZI4 A0A232EZ04 K7INE2 A0A0P5E6X7 A0A0P5D1I1 A0A224Z4S9 A0A1S3JWF3 A0A067RIW3 A0A0P5AGP6 A0A0P5NG54 A0A194PDE3 A0A293MSK5 A0A087UJK6 A0A194RHH9 A0A087SZR9 A0A310S8J2 A0A1E1X7M4 A0A0K2TDT5 A0A0N8B201 A0A0N8A4H0 A0A0P4Z373 A0A0P5T7C5 A0A0P5II29 A0A2R5LA64 A0A0N8CJA7 A0A023GE63 A0A0P4Z993 A0A088AI46 A0A154PE44 A0A1A9WIQ9 A0A0P4Z6R4 A0A087T9D3 Q293F9 H9IUJ7 A0A2P6L6H6 W8B744 E2C7A6 B4MYD7 T1J119 A0A0P6DBF2 A0A1A9Y0M4 A0A212EKK6 A0A0N8EJE9 A0A2T7PMP6 E0VAE8 A0A1I8J6K1 A0A1S3INW9
Pubmed
EMBL
KZ149969
PZC76099.1
ODYU01006554
SOQ48496.1
KQ461198
KPJ05995.1
+ More
GDQN01006111 GDQN01002640 JAT84943.1 JAT88414.1 AGBW02008939 OWR52121.1 RSAL01000048 RVE50485.1 KQ459185 KPJ03392.1 UFQS01000091 UFQT01000091 SSW99174.1 SSX19556.1 CVRI01000051 CRK99499.1 GFDF01007418 JAV06666.1 KQ971377 EEZ97453.2 KYB25101.1 GBYB01007728 JAG77495.1 GL440813 EFN65393.1 CVRI01000057 CRL02312.1 CVRI01000038 CRK94129.1 CRK94126.1 QOIP01000006 RLU22213.1 NNAY01003483 OXU19349.1 NEVH01020850 PNF21288.1 ADTU01014843 GL452135 EFN77827.1 KB631673 ERL85279.1 APGK01047262 KB741077 ENN74298.1 KQ435710 KOX79786.1 CRK94128.1 KQ979568 KYN20906.1 CRK94127.1 KQ982314 KYQ57887.1 GL888818 EGI57914.1 KQ981864 KYN34231.1 KQ976780 KYN08349.1 GL768221 EFZ11724.1 KQ976662 KYM78572.1 EF710645 ACE75156.1 PYGN01000157 PSN52584.1 AJVK01009496 AMQM01000267 KB095811 ESO12934.1 IACF01002309 LAB67968.1 GEGO01004964 JAR90440.1 GEFH01000809 JAP67772.1 GDIP01089951 JAM13764.1 GDIQ01068403 JAN26334.1 GFPF01011892 MAA23038.1 NNAY01001559 OXU23601.1 GDIP01146239 JAJ77163.1 GDIP01162514 JAJ60888.1 GFPF01011893 MAA23039.1 KK852446 KDR23737.1 GDIP01199386 JAJ24016.1 GDIQ01142620 JAL09106.1 KQ459606 KPI91267.1 GFWV01018351 MAA43079.1 KK120113 KFM77545.1 KQ460205 KPJ16894.1 KK112715 KFM58358.1 KQ770716 OAD52621.1 GFAC01003928 JAT95260.1 HACA01006644 CDW24005.1 GDIQ01220651 JAK31074.1 GDIP01183377 JAJ40025.1 GDIP01230369 GDIP01219074 JAJ04328.1 GDIP01136485 JAL67229.1 GDIQ01237904 JAK13821.1 GGLE01002278 MBY06404.1 GDIP01126191 JAL77523.1 GBBM01003289 JAC32129.1 GDIP01219072 JAJ04330.1 KQ434886 KZC10146.1 GDIP01217287 JAJ06115.1 KK114112 KFM61722.1 CM000071 EAL24652.3 BABH01000949 MWRG01001453 PRD34179.1 GAMC01009565 JAB96990.1 GL453340 EFN76219.1 CH963894 EDW77126.2 JH431752 GDIQ01079731 JAN15006.1 AGBW02014254 OWR42013.1 GDIQ01020876 JAN73861.1 PZQS01000003 PVD34691.1 DS235005 EEB10354.1 NIVC01000084 PAA91526.1
GDQN01006111 GDQN01002640 JAT84943.1 JAT88414.1 AGBW02008939 OWR52121.1 RSAL01000048 RVE50485.1 KQ459185 KPJ03392.1 UFQS01000091 UFQT01000091 SSW99174.1 SSX19556.1 CVRI01000051 CRK99499.1 GFDF01007418 JAV06666.1 KQ971377 EEZ97453.2 KYB25101.1 GBYB01007728 JAG77495.1 GL440813 EFN65393.1 CVRI01000057 CRL02312.1 CVRI01000038 CRK94129.1 CRK94126.1 QOIP01000006 RLU22213.1 NNAY01003483 OXU19349.1 NEVH01020850 PNF21288.1 ADTU01014843 GL452135 EFN77827.1 KB631673 ERL85279.1 APGK01047262 KB741077 ENN74298.1 KQ435710 KOX79786.1 CRK94128.1 KQ979568 KYN20906.1 CRK94127.1 KQ982314 KYQ57887.1 GL888818 EGI57914.1 KQ981864 KYN34231.1 KQ976780 KYN08349.1 GL768221 EFZ11724.1 KQ976662 KYM78572.1 EF710645 ACE75156.1 PYGN01000157 PSN52584.1 AJVK01009496 AMQM01000267 KB095811 ESO12934.1 IACF01002309 LAB67968.1 GEGO01004964 JAR90440.1 GEFH01000809 JAP67772.1 GDIP01089951 JAM13764.1 GDIQ01068403 JAN26334.1 GFPF01011892 MAA23038.1 NNAY01001559 OXU23601.1 GDIP01146239 JAJ77163.1 GDIP01162514 JAJ60888.1 GFPF01011893 MAA23039.1 KK852446 KDR23737.1 GDIP01199386 JAJ24016.1 GDIQ01142620 JAL09106.1 KQ459606 KPI91267.1 GFWV01018351 MAA43079.1 KK120113 KFM77545.1 KQ460205 KPJ16894.1 KK112715 KFM58358.1 KQ770716 OAD52621.1 GFAC01003928 JAT95260.1 HACA01006644 CDW24005.1 GDIQ01220651 JAK31074.1 GDIP01183377 JAJ40025.1 GDIP01230369 GDIP01219074 JAJ04328.1 GDIP01136485 JAL67229.1 GDIQ01237904 JAK13821.1 GGLE01002278 MBY06404.1 GDIP01126191 JAL77523.1 GBBM01003289 JAC32129.1 GDIP01219072 JAJ04330.1 KQ434886 KZC10146.1 GDIP01217287 JAJ06115.1 KK114112 KFM61722.1 CM000071 EAL24652.3 BABH01000949 MWRG01001453 PRD34179.1 GAMC01009565 JAB96990.1 GL453340 EFN76219.1 CH963894 EDW77126.2 JH431752 GDIQ01079731 JAN15006.1 AGBW02014254 OWR42013.1 GDIQ01020876 JAN73861.1 PZQS01000003 PVD34691.1 DS235005 EEB10354.1 NIVC01000084 PAA91526.1
Proteomes
UP000053240
UP000007151
UP000283053
UP000053268
UP000183832
UP000007266
+ More
UP000005203 UP000000311 UP000192223 UP000279307 UP000215335 UP000235965 UP000005205 UP000008237 UP000030742 UP000019118 UP000053105 UP000078492 UP000075809 UP000007755 UP000078541 UP000078542 UP000078540 UP000245037 UP000092462 UP000015101 UP000085678 UP000002358 UP000027135 UP000054359 UP000076502 UP000091820 UP000001819 UP000005204 UP000007798 UP000092443 UP000245119 UP000009046 UP000095280 UP000215902
UP000005203 UP000000311 UP000192223 UP000279307 UP000215335 UP000235965 UP000005205 UP000008237 UP000030742 UP000019118 UP000053105 UP000078492 UP000075809 UP000007755 UP000078541 UP000078542 UP000078540 UP000245037 UP000092462 UP000015101 UP000085678 UP000002358 UP000027135 UP000054359 UP000076502 UP000091820 UP000001819 UP000005204 UP000007798 UP000092443 UP000245119 UP000009046 UP000095280 UP000215902
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BM12
A0A2H1W6C2
A0A194QM12
A0A1E1WN46
A0A212FEF8
A0A3S2NLB0
+ More
A0A194QD56 A0A336LNJ2 A0A1J1IH02 A0A1L8DJN8 D6X3Q2 A0A139WB82 A0A088AFL2 A0A0C9Q1F8 E2AMG9 A0A1J1IQ76 A0A1J1I6T3 A0A1W4X5F8 A0A1J1I5D3 A0A3L8DPQ4 A0A232ELW2 A0A2J7PY70 A0A158NGE2 E2C2H1 U4TUP7 N6T994 A0A1W4WUR5 A0A0M9AAF7 A0A1J1I1C2 A0A195E733 A0A1J1I1A1 A0A151XC17 F4X6Y2 A0A195F1H6 A0A151IQB6 E9J630 A0A195B2Q5 B7S8D1 A0A2P8Z7V4 A0A1B0CZ55 T1FPK8 A0A2P2I1R7 A0A1S3HS51 A0A1S3HPA4 K7IZ11 A0A147BI19 A0A131XN80 A0A0N8CX87 A0A0P6E398 A0A224YZI4 A0A232EZ04 K7INE2 A0A0P5E6X7 A0A0P5D1I1 A0A224Z4S9 A0A1S3JWF3 A0A067RIW3 A0A0P5AGP6 A0A0P5NG54 A0A194PDE3 A0A293MSK5 A0A087UJK6 A0A194RHH9 A0A087SZR9 A0A310S8J2 A0A1E1X7M4 A0A0K2TDT5 A0A0N8B201 A0A0N8A4H0 A0A0P4Z373 A0A0P5T7C5 A0A0P5II29 A0A2R5LA64 A0A0N8CJA7 A0A023GE63 A0A0P4Z993 A0A088AI46 A0A154PE44 A0A1A9WIQ9 A0A0P4Z6R4 A0A087T9D3 Q293F9 H9IUJ7 A0A2P6L6H6 W8B744 E2C7A6 B4MYD7 T1J119 A0A0P6DBF2 A0A1A9Y0M4 A0A212EKK6 A0A0N8EJE9 A0A2T7PMP6 E0VAE8 A0A1I8J6K1 A0A1S3INW9
A0A194QD56 A0A336LNJ2 A0A1J1IH02 A0A1L8DJN8 D6X3Q2 A0A139WB82 A0A088AFL2 A0A0C9Q1F8 E2AMG9 A0A1J1IQ76 A0A1J1I6T3 A0A1W4X5F8 A0A1J1I5D3 A0A3L8DPQ4 A0A232ELW2 A0A2J7PY70 A0A158NGE2 E2C2H1 U4TUP7 N6T994 A0A1W4WUR5 A0A0M9AAF7 A0A1J1I1C2 A0A195E733 A0A1J1I1A1 A0A151XC17 F4X6Y2 A0A195F1H6 A0A151IQB6 E9J630 A0A195B2Q5 B7S8D1 A0A2P8Z7V4 A0A1B0CZ55 T1FPK8 A0A2P2I1R7 A0A1S3HS51 A0A1S3HPA4 K7IZ11 A0A147BI19 A0A131XN80 A0A0N8CX87 A0A0P6E398 A0A224YZI4 A0A232EZ04 K7INE2 A0A0P5E6X7 A0A0P5D1I1 A0A224Z4S9 A0A1S3JWF3 A0A067RIW3 A0A0P5AGP6 A0A0P5NG54 A0A194PDE3 A0A293MSK5 A0A087UJK6 A0A194RHH9 A0A087SZR9 A0A310S8J2 A0A1E1X7M4 A0A0K2TDT5 A0A0N8B201 A0A0N8A4H0 A0A0P4Z373 A0A0P5T7C5 A0A0P5II29 A0A2R5LA64 A0A0N8CJA7 A0A023GE63 A0A0P4Z993 A0A088AI46 A0A154PE44 A0A1A9WIQ9 A0A0P4Z6R4 A0A087T9D3 Q293F9 H9IUJ7 A0A2P6L6H6 W8B744 E2C7A6 B4MYD7 T1J119 A0A0P6DBF2 A0A1A9Y0M4 A0A212EKK6 A0A0N8EJE9 A0A2T7PMP6 E0VAE8 A0A1I8J6K1 A0A1S3INW9
PDB
4XPG
E-value=2.03262e-88,
Score=833
Ontologies
GO
PANTHER
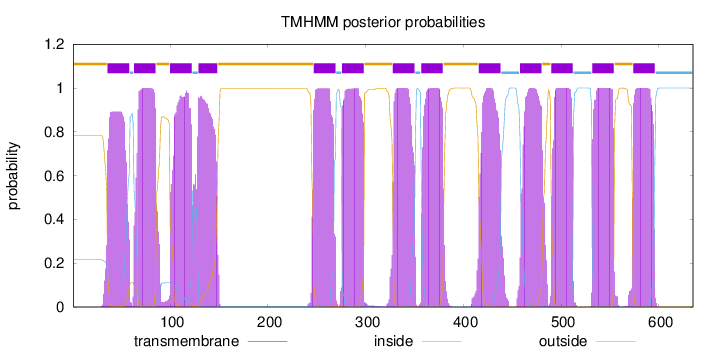
Topology
Length:
635
Number of predicted TMHs:
13
Exp number of AAs in TMHs:
285.51289
Exp number, first 60 AAs:
18.89259
Total prob of N-in:
0.21621
POSSIBLE N-term signal
sequence
outside
1 - 35
TMhelix
36 - 58
inside
59 - 62
TMhelix
63 - 85
outside
86 - 99
TMhelix
100 - 122
inside
123 - 128
TMhelix
129 - 148
outside
149 - 246
TMhelix
247 - 269
inside
270 - 275
TMhelix
276 - 298
outside
299 - 327
TMhelix
328 - 350
inside
351 - 356
TMhelix
357 - 379
outside
380 - 415
TMhelix
416 - 438
inside
439 - 457
TMhelix
458 - 480
outside
481 - 489
TMhelix
490 - 512
inside
513 - 531
TMhelix
532 - 554
outside
555 - 573
TMhelix
574 - 596
inside
597 - 635
Population Genetic Test Statistics
Pi
21.213558
Theta
188.546266
Tajima's D
0.640369
CLR
1.070366
CSRT
0.553272336383181
Interpretation
Uncertain
