Gene
KWMTBOMO04862
Annotation
PREDICTED:_allantoicase-like_[Amyelois_transitella]
Full name
Allantoicase
+ More
Probable allantoicase
Probable allantoicase
Alternative Name
Allantoate amidinohydrolase
Location in the cell
Cytoplasmic Reliability : 1.058 Nuclear Reliability : 1.543
Sequence
CDS
ATGGCTGATGGGTGGGAGACGGCGAGAAGGGCGGATAGGCCCGAGATCATCGAAGAGAATGATGATGGAACTCTGAGGTTTTCAGGCAACGAGTGGGCAGTCTACAAATTAGGTTTCGCTGGTACAATAAGAAGAGTGTGCATTGACACAACACATTTCAAAGGCAATTATCCAGATCATATCAGGATTGAAGGCTGCAATTTGGGACAGAATGATTGGGATAATCAATCCAATAACGAATGTAATTGGAGAAGCATATTGGTACCGAGCAAGGTAAGAAATTGA
Protein
MADGWETARRADRPEIIEENDDGTLRFSGNEWAVYKLGFAGTIRRVCIDTTHFKGNYPDHIRIEGCNLGQNDWDNQSNNECNWRSILVPSKVRN
Summary
Description
Utilization of purines as secondary nitrogen sources, when primary sources are limiting.
Catalytic Activity
allantoate + H2O = (S)-ureidoglycolate + urea
Similarity
Belongs to the allantoicase family.
Keywords
Alternative splicing
Complete proteome
Hydrolase
Purine metabolism
Reference proteome
Feature
chain Allantoicase
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2H1V906
A0A3S2L6D3
A0A2W1BIP5
A0A2A4JAL9
S4NN49
A0A194QKS2
+ More
A0A194QIW1 A0A212EIY9 H9JET3 K1QWU9 A0A0C9RH93 Q7Z086 W4YU35 Q6DGA6-2 B2GQF5 Q6DGA6 A0A1W4Z5J6 A0A0P7V9D1 A0A1W4ZE74 A0A0C9RXS2 A0A3P8V250 A0A3B3RE78 A0A3P9KUW0 A0A3P9HQE9 H2MV89 A0A2P8XJG3 A0A3B1IGN1 F7ERQ3 A0A1Y1KFG4 A0A3Q4GBL4 A0A1Y1KIF9 A0A3Q4GGX1 A0A3N0XI67 A0A3B4FA36 A0A2R2MNJ6 A0A3P8PXU7 A0A3Q2X649 A0A3P8PYE4 A0A3P9CBE1 A0A1Y1KFR9 A7SUH3 A0A3R7LZH5 A0A3Q1DF69 A0A3Q1C888 A0A3P8XYZ4 A0A3Q3XDN3 K7F542 A0A1A8VG99 A0A2D0Q2X1 A0A3P8TUE1 A0A3Q1FPR4 W5NKW8 A0A3Q1GUC1 A0A3Q3GB37 A0A2D0PYY5 A0A2D0Q0A1 A0A3B4EED3 A0A3B4EEC1 A0A3L7HCA9 H2SPJ4 A0A3B3DE46 W5NKW7 A0A3Q3N4M2 A0A0P4WMJ2 A0A146YA36 I3KRF6 S4W3N7 A0A060VYL2 A0A0P4WF94 A0A1U7R8Y4 A0A3B3THJ0 G3I3C7 H0WGE6 A0A3B5MPR7 A0A3B5QYG2 D2I2B1 A0A1S3MBD9 A0A2K6KYH6 A0A096NIM5 A0A2K5YKA3 A0A087YE11 F6VB05 G1MJF8 A0A146NUD1 A0A3B4UTX7 A0A2K6BJ99 A0A384BN17 A0A182H1C0 A0A146NDC2 A0A2K5LJL9 G1TAW4 G8F4Y6 F6TE28 Q4R964 A0A3Q3L3Q6 A0A146YA87 A0A2K6BJC1 A0A0T6BFF9 A0A2K6PK05 A0A3Q2ZYG9
A0A194QIW1 A0A212EIY9 H9JET3 K1QWU9 A0A0C9RH93 Q7Z086 W4YU35 Q6DGA6-2 B2GQF5 Q6DGA6 A0A1W4Z5J6 A0A0P7V9D1 A0A1W4ZE74 A0A0C9RXS2 A0A3P8V250 A0A3B3RE78 A0A3P9KUW0 A0A3P9HQE9 H2MV89 A0A2P8XJG3 A0A3B1IGN1 F7ERQ3 A0A1Y1KFG4 A0A3Q4GBL4 A0A1Y1KIF9 A0A3Q4GGX1 A0A3N0XI67 A0A3B4FA36 A0A2R2MNJ6 A0A3P8PXU7 A0A3Q2X649 A0A3P8PYE4 A0A3P9CBE1 A0A1Y1KFR9 A7SUH3 A0A3R7LZH5 A0A3Q1DF69 A0A3Q1C888 A0A3P8XYZ4 A0A3Q3XDN3 K7F542 A0A1A8VG99 A0A2D0Q2X1 A0A3P8TUE1 A0A3Q1FPR4 W5NKW8 A0A3Q1GUC1 A0A3Q3GB37 A0A2D0PYY5 A0A2D0Q0A1 A0A3B4EED3 A0A3B4EEC1 A0A3L7HCA9 H2SPJ4 A0A3B3DE46 W5NKW7 A0A3Q3N4M2 A0A0P4WMJ2 A0A146YA36 I3KRF6 S4W3N7 A0A060VYL2 A0A0P4WF94 A0A1U7R8Y4 A0A3B3THJ0 G3I3C7 H0WGE6 A0A3B5MPR7 A0A3B5QYG2 D2I2B1 A0A1S3MBD9 A0A2K6KYH6 A0A096NIM5 A0A2K5YKA3 A0A087YE11 F6VB05 G1MJF8 A0A146NUD1 A0A3B4UTX7 A0A2K6BJ99 A0A384BN17 A0A182H1C0 A0A146NDC2 A0A2K5LJL9 G1TAW4 G8F4Y6 F6TE28 Q4R964 A0A3Q3L3Q6 A0A146YA87 A0A2K6BJC1 A0A0T6BFF9 A0A2K6PK05 A0A3Q2ZYG9
EC Number
3.5.3.4
Pubmed
EMBL
ODYU01001289
SOQ37297.1
RSAL01000121
RVE46686.1
KZ150019
PZC74932.1
+ More
NWSH01002108 PCG69157.1 GAIX01014086 JAA78474.1 KQ461198 KPJ05994.1 KQ459185 KPJ03391.1 AGBW02014553 OWR41445.1 BABH01023327 BABH01023328 BABH01023329 JH817383 EKC35614.1 GBYB01007670 JAG77437.1 AY269784 AAP41914.1 AAGJ04010450 BX276180 BC076445 BC164715 AAI64715.1 JARO02000371 KPP78802.1 GBYB01014020 JAG83787.1 PYGN01001921 PSN32141.1 GEZM01085131 GEZM01085128 JAV60199.1 GEZM01085132 GEZM01085129 JAV60201.1 RJVU01072567 ROI33889.1 GEZM01085130 GEZM01085127 JAV60204.1 DS469813 EDO32648.1 QCYY01002635 ROT68777.1 AGCU01012893 HAEJ01019217 SBS59674.1 AHAT01032617 RAZU01000241 RLQ63260.1 GDRN01061079 JAI65249.1 GCES01035868 JAR50455.1 AERX01016628 KF137576 AGO86379.1 FR904338 CDQ59942.1 GDRN01061077 JAI65250.1 JH001181 EGW10652.1 AAQR03147187 GL194142 EFB16901.1 AHZZ02004863 AYCK01010430 ACTA01160405 GCES01151207 JAQ35115.1 JXUM01102942 KQ564765 KXJ71772.1 GCES01157276 JAQ29046.1 JH331173 EHH62350.1 JSUE03010094 JH290552 EHH25475.1 AB168232 GCES01035871 JAR50452.1 LJIG01001039 KRT85901.1
NWSH01002108 PCG69157.1 GAIX01014086 JAA78474.1 KQ461198 KPJ05994.1 KQ459185 KPJ03391.1 AGBW02014553 OWR41445.1 BABH01023327 BABH01023328 BABH01023329 JH817383 EKC35614.1 GBYB01007670 JAG77437.1 AY269784 AAP41914.1 AAGJ04010450 BX276180 BC076445 BC164715 AAI64715.1 JARO02000371 KPP78802.1 GBYB01014020 JAG83787.1 PYGN01001921 PSN32141.1 GEZM01085131 GEZM01085128 JAV60199.1 GEZM01085132 GEZM01085129 JAV60201.1 RJVU01072567 ROI33889.1 GEZM01085130 GEZM01085127 JAV60204.1 DS469813 EDO32648.1 QCYY01002635 ROT68777.1 AGCU01012893 HAEJ01019217 SBS59674.1 AHAT01032617 RAZU01000241 RLQ63260.1 GDRN01061079 JAI65249.1 GCES01035868 JAR50455.1 AERX01016628 KF137576 AGO86379.1 FR904338 CDQ59942.1 GDRN01061077 JAI65250.1 JH001181 EGW10652.1 AAQR03147187 GL194142 EFB16901.1 AHZZ02004863 AYCK01010430 ACTA01160405 GCES01151207 JAQ35115.1 JXUM01102942 KQ564765 KXJ71772.1 GCES01157276 JAQ29046.1 JH331173 EHH62350.1 JSUE03010094 JH290552 EHH25475.1 AB168232 GCES01035871 JAR50452.1 LJIG01001039 KRT85901.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
UP000005204
+ More
UP000005408 UP000007110 UP000000437 UP000192224 UP000034805 UP000265120 UP000261540 UP000265180 UP000265200 UP000001038 UP000245037 UP000018467 UP000002280 UP000261580 UP000261460 UP000085678 UP000265100 UP000264840 UP000265160 UP000001593 UP000283509 UP000257160 UP000265140 UP000261620 UP000007267 UP000221080 UP000265080 UP000257200 UP000018468 UP000261660 UP000261440 UP000273346 UP000005226 UP000261560 UP000005207 UP000193380 UP000189706 UP000261500 UP000001075 UP000005225 UP000261380 UP000002852 UP000087266 UP000233180 UP000028761 UP000233140 UP000028760 UP000002279 UP000008912 UP000261420 UP000233120 UP000261680 UP000069940 UP000249989 UP000233060 UP000001811 UP000009130 UP000006718 UP000233100 UP000261640 UP000233200 UP000264800
UP000005408 UP000007110 UP000000437 UP000192224 UP000034805 UP000265120 UP000261540 UP000265180 UP000265200 UP000001038 UP000245037 UP000018467 UP000002280 UP000261580 UP000261460 UP000085678 UP000265100 UP000264840 UP000265160 UP000001593 UP000283509 UP000257160 UP000265140 UP000261620 UP000007267 UP000221080 UP000265080 UP000257200 UP000018468 UP000261660 UP000261440 UP000273346 UP000005226 UP000261560 UP000005207 UP000193380 UP000189706 UP000261500 UP000001075 UP000005225 UP000261380 UP000002852 UP000087266 UP000233180 UP000028761 UP000233140 UP000028760 UP000002279 UP000008912 UP000261420 UP000233120 UP000261680 UP000069940 UP000249989 UP000233060 UP000001811 UP000009130 UP000006718 UP000233100 UP000261640 UP000233200 UP000264800
Interpro
Gene 3D
ProteinModelPortal
A0A2H1V906
A0A3S2L6D3
A0A2W1BIP5
A0A2A4JAL9
S4NN49
A0A194QKS2
+ More
A0A194QIW1 A0A212EIY9 H9JET3 K1QWU9 A0A0C9RH93 Q7Z086 W4YU35 Q6DGA6-2 B2GQF5 Q6DGA6 A0A1W4Z5J6 A0A0P7V9D1 A0A1W4ZE74 A0A0C9RXS2 A0A3P8V250 A0A3B3RE78 A0A3P9KUW0 A0A3P9HQE9 H2MV89 A0A2P8XJG3 A0A3B1IGN1 F7ERQ3 A0A1Y1KFG4 A0A3Q4GBL4 A0A1Y1KIF9 A0A3Q4GGX1 A0A3N0XI67 A0A3B4FA36 A0A2R2MNJ6 A0A3P8PXU7 A0A3Q2X649 A0A3P8PYE4 A0A3P9CBE1 A0A1Y1KFR9 A7SUH3 A0A3R7LZH5 A0A3Q1DF69 A0A3Q1C888 A0A3P8XYZ4 A0A3Q3XDN3 K7F542 A0A1A8VG99 A0A2D0Q2X1 A0A3P8TUE1 A0A3Q1FPR4 W5NKW8 A0A3Q1GUC1 A0A3Q3GB37 A0A2D0PYY5 A0A2D0Q0A1 A0A3B4EED3 A0A3B4EEC1 A0A3L7HCA9 H2SPJ4 A0A3B3DE46 W5NKW7 A0A3Q3N4M2 A0A0P4WMJ2 A0A146YA36 I3KRF6 S4W3N7 A0A060VYL2 A0A0P4WF94 A0A1U7R8Y4 A0A3B3THJ0 G3I3C7 H0WGE6 A0A3B5MPR7 A0A3B5QYG2 D2I2B1 A0A1S3MBD9 A0A2K6KYH6 A0A096NIM5 A0A2K5YKA3 A0A087YE11 F6VB05 G1MJF8 A0A146NUD1 A0A3B4UTX7 A0A2K6BJ99 A0A384BN17 A0A182H1C0 A0A146NDC2 A0A2K5LJL9 G1TAW4 G8F4Y6 F6TE28 Q4R964 A0A3Q3L3Q6 A0A146YA87 A0A2K6BJC1 A0A0T6BFF9 A0A2K6PK05 A0A3Q2ZYG9
A0A194QIW1 A0A212EIY9 H9JET3 K1QWU9 A0A0C9RH93 Q7Z086 W4YU35 Q6DGA6-2 B2GQF5 Q6DGA6 A0A1W4Z5J6 A0A0P7V9D1 A0A1W4ZE74 A0A0C9RXS2 A0A3P8V250 A0A3B3RE78 A0A3P9KUW0 A0A3P9HQE9 H2MV89 A0A2P8XJG3 A0A3B1IGN1 F7ERQ3 A0A1Y1KFG4 A0A3Q4GBL4 A0A1Y1KIF9 A0A3Q4GGX1 A0A3N0XI67 A0A3B4FA36 A0A2R2MNJ6 A0A3P8PXU7 A0A3Q2X649 A0A3P8PYE4 A0A3P9CBE1 A0A1Y1KFR9 A7SUH3 A0A3R7LZH5 A0A3Q1DF69 A0A3Q1C888 A0A3P8XYZ4 A0A3Q3XDN3 K7F542 A0A1A8VG99 A0A2D0Q2X1 A0A3P8TUE1 A0A3Q1FPR4 W5NKW8 A0A3Q1GUC1 A0A3Q3GB37 A0A2D0PYY5 A0A2D0Q0A1 A0A3B4EED3 A0A3B4EEC1 A0A3L7HCA9 H2SPJ4 A0A3B3DE46 W5NKW7 A0A3Q3N4M2 A0A0P4WMJ2 A0A146YA36 I3KRF6 S4W3N7 A0A060VYL2 A0A0P4WF94 A0A1U7R8Y4 A0A3B3THJ0 G3I3C7 H0WGE6 A0A3B5MPR7 A0A3B5QYG2 D2I2B1 A0A1S3MBD9 A0A2K6KYH6 A0A096NIM5 A0A2K5YKA3 A0A087YE11 F6VB05 G1MJF8 A0A146NUD1 A0A3B4UTX7 A0A2K6BJ99 A0A384BN17 A0A182H1C0 A0A146NDC2 A0A2K5LJL9 G1TAW4 G8F4Y6 F6TE28 Q4R964 A0A3Q3L3Q6 A0A146YA87 A0A2K6BJC1 A0A0T6BFF9 A0A2K6PK05 A0A3Q2ZYG9
PDB
1O59
E-value=1.17485e-06,
Score=118
Ontologies
PATHWAY
GO
PANTHER
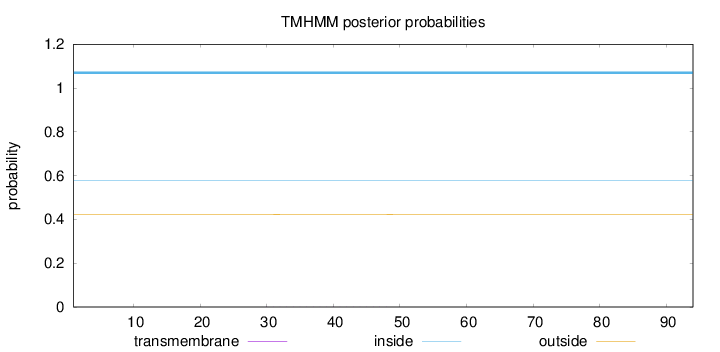
Topology
Length:
94
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00727
Exp number, first 60 AAs:
0.00727
Total prob of N-in:
0.57760
inside
1 - 94
Population Genetic Test Statistics
Pi
104.775061
Theta
82.8106
Tajima's D
0.830265
CLR
0
CSRT
0.619869006549672
Interpretation
Uncertain
