Gene
KWMTBOMO04858
Pre Gene Modal
BGIBMGA008045
Annotation
PREDICTED:_acid_sphingomyelinase-like_phosphodiesterase_3a_isoform_X2_[Papilio_xuthus]
Full name
Sphingomyelinase phosphodiesterase
+ More
Acid sphingomyelinase-like phosphodiesterase 3b
Acid sphingomyelinase-like phosphodiesterase
Acid sphingomyelinase-like phosphodiesterase 3b
Acid sphingomyelinase-like phosphodiesterase
Location in the cell
PlasmaMembrane Reliability : 2.224
Sequence
CDS
ATGCTGACCCTAAAAGATTTTTCTACTGATCTGTACTTTATAGTGCAAATGGCTGATATGACACTCTCCTGCTCTGTGTCACAAGGACAATATGAAATAAATTGTCGGAGATTAGCCGATCGTGGTAGTTCAAGATATCTCAGGGCTCTGGGGAAATATGGGGACTACGCCTGTGACAGCTCTTTGGAGCTCATCGAATCCGCTGCAAAGTACATGAGAACCCGGCACTCGGATAACGTAGAATTCGTATTGTGGACAGGGGACATAGTCACTTCCCACTACACGCTAAGCGAAGACGACCGTCTGAACGCTATCCGTAACATGACGTTCCTATTGAGTAGGACCTTCAGTTCTCACTTCGTGTTTCCTGTTCTCGGGGACTTGGACCCCAAGCCATCCGAAACTGTCACTACTCTATGGATGCAATGGCTACCTTTGGAAGCTTTAGAGACCTTCGCTAATGGTGGGTACTACACGATTGAGCAGAAACAGAACAAGCTGAGGCTTGTGATCCTGAACACGAACTTGTTCGGGATTCGTGAGCACAATACCCCAGAGTCAAGTCTGCAGTGGGAATGGCTCAGCTCCGTGTTCAACAAGGCGGACAAGGCTAACGAAATGATATACATCATAGGCCACACGGCGCCCGGGACGGAATATCGCCATGGGCACGAGATGGCGGTCGGCGCCGTGGAGAAATACATCAGGATGGTCCGTCAACACGCTCGCCGGATTGCCGGACAGTTCTTCGGGCACCTTCACACTGACACCTTCAGGATCTTCTATGACAATGAGCGTCCAGTATCTTGGGCACTGCTGGCGCCGTCAGTAACACCTCATCGGTACCCTAATGATGCCTCAAACCCTGGCCTGAGGCTGTATAAATTTGACACCCACAATGGAAAGGTATTAGATTACACCCAGTACTACTTAGATCTGAAAACAGCGAACCGCAATATAGAGTCTGCTCAGTGGACGGTAGAGTACAACGTGACTCTGTATTACGGACTCCACGATATCACAGCCACTTCTCTACATAACCTTGCCGATAGGATACGAACGGGAACCACTCACGATAATTCTGTTCTTAAGAGGTATCTGAGGGCGTACAAAGTAAAATTTGAGGAGTCAGAAAACTGCGACAGCGCGTGTGCTCACCAGCATTACTGCTCGATCACCTGCCTCGACCTTCCCGCCTACCAGCAGTGCCTAGACTCTGCCAGCAGTGCCCTGGCTACTGCGGCAGATTCTGCCGCCATCGAAACTTTCCCGCTCAAAGCCGCCATTGCACTGACCATTTTCGCCGCTGGAGTCTCGTTTTGA
Protein
MLTLKDFSTDLYFIVQMADMTLSCSVSQGQYEINCRRLADRGSSRYLRALGKYGDYACDSSLELIESAAKYMRTRHSDNVEFVLWTGDIVTSHYTLSEDDRLNAIRNMTFLLSRTFSSHFVFPVLGDLDPKPSETVTTLWMQWLPLEALETFANGGYYTIEQKQNKLRLVILNTNLFGIREHNTPESSLQWEWLSSVFNKADKANEMIYIIGHTAPGTEYRHGHEMAVGAVEKYIRMVRQHARRIAGQFFGHLHTDTFRIFYDNERPVSWALLAPSVTPHRYPNDASNPGLRLYKFDTHNGKVLDYTQYYLDLKTANRNIESAQWTVEYNVTLYYGLHDITATSLHNLADRIRTGTTHDNSVLKRYLRAYKVKFEESENCDSACAHQHYCSITCLDLPAYQQCLDSASSALATAADSAAIETFPLKAAIALTIFAAGVSF
Summary
Description
Lipid-modulating phosphodiesterase (PubMed:26095358). Active on the surface of macrophages and dendritic cells and strongly influences macrophage lipid composition and membrane fluidity. Acts as a negative regulator of Toll-like receptor signaling (By similarity). Has in vitro phosphodiesterase activity, but the physiological substrate is unknown (PubMed:26095358). Lacks activity with phosphocholine-containing lipids, but can cleave CDP-choline, and can release phosphate from ATP and ADP (in vitro) (By similarity).
Cofactor
Zn(2+)
Subunit
Interacts with TLR4, TLR7, TLR8 and TLR9.
Similarity
Belongs to the acid sphingomyelinase family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
Glycosidase
GPI-anchor
Hydrolase
Immunity
Inflammatory response
Innate immunity
Lipid degradation
Lipid metabolism
Lipoprotein
Membrane
Metal-binding
Polymorphism
Reference proteome
Secreted
Signal
Zinc
Feature
chain Acid sphingomyelinase-like phosphodiesterase 3b
splice variant In isoform 2.
sequence variant In dbSNP:rs34560878.
splice variant In isoform 2.
sequence variant In dbSNP:rs34560878.
Uniprot
H9JEU8
A0A2W1BKX4
A0A2H1W9F8
A0A3S2N4T6
A0A194QR38
A0A194QCX4
+ More
A0A212EIY4 A0A2A4JBL4 A0A2H1VY23 A0A194QIT8 A0A212ES85 A0A139WAW8 A0A067RJ29 A0A0N1IQ10 A0A1W4X5I9 A0A2S2QMJ5 E0VZL8 A0A154PQH5 Q172E7 A0A2H8U085 A0A195F186 A0A195E6T8 A0A151IQE7 F4X6F8 A0A3B0JZE6 E2AMG6 A0A195B2Q9 A0A151XBZ8 B4J2N4 A0A026VYW8 B4PDW7 B4N490 A0A2A3EAU8 A0A088AER8 B3MA63 B4LG96 B4HAE2 Q2LZA0 A0A0Q9WI47 A0A158NGE5 A0A0M4F0F5 A0A1B6FNW2 A0A0M9A977 A0A336LRH4 A0A0C9QK20 A0A1D2NBQ4 A0A2P8XVA8 A0A0P4Y4T5 A0A0N8A3F6 A0A0P6DYQ7 A0A0P6HED9 T1HWJ8 A0A0P5DS43 A0A0P6C7J9 A0A0P5JPY0 H2YX51 Q92485 A0A2R9AWT9 J9NYQ3 A0A340XCA0 A0A383Z5Z0 A0A2J8JP43 B4DW34 A0A1U7U7J5 A0A3Q2NN50 S7PQU8 F7C9L5 G3RG15 K9IL01 A0A3Q7TVF7 L8IID3 G1QCT9 A6QQN6 A0A2B4RQL5 A0A3N0Y311 A0A3Q3G0E0 K7GCP6 A0A3Q2EAZ6 A0A0N7ZAS1 H2N8B7 A0A2Y9SVS0 A0A1U8DZB1 I3M4A5 G1T3N0 W5M6L3 H0XDL1
A0A212EIY4 A0A2A4JBL4 A0A2H1VY23 A0A194QIT8 A0A212ES85 A0A139WAW8 A0A067RJ29 A0A0N1IQ10 A0A1W4X5I9 A0A2S2QMJ5 E0VZL8 A0A154PQH5 Q172E7 A0A2H8U085 A0A195F186 A0A195E6T8 A0A151IQE7 F4X6F8 A0A3B0JZE6 E2AMG6 A0A195B2Q9 A0A151XBZ8 B4J2N4 A0A026VYW8 B4PDW7 B4N490 A0A2A3EAU8 A0A088AER8 B3MA63 B4LG96 B4HAE2 Q2LZA0 A0A0Q9WI47 A0A158NGE5 A0A0M4F0F5 A0A1B6FNW2 A0A0M9A977 A0A336LRH4 A0A0C9QK20 A0A1D2NBQ4 A0A2P8XVA8 A0A0P4Y4T5 A0A0N8A3F6 A0A0P6DYQ7 A0A0P6HED9 T1HWJ8 A0A0P5DS43 A0A0P6C7J9 A0A0P5JPY0 H2YX51 Q92485 A0A2R9AWT9 J9NYQ3 A0A340XCA0 A0A383Z5Z0 A0A2J8JP43 B4DW34 A0A1U7U7J5 A0A3Q2NN50 S7PQU8 F7C9L5 G3RG15 K9IL01 A0A3Q7TVF7 L8IID3 G1QCT9 A6QQN6 A0A2B4RQL5 A0A3N0Y311 A0A3Q3G0E0 K7GCP6 A0A3Q2EAZ6 A0A0N7ZAS1 H2N8B7 A0A2Y9SVS0 A0A1U8DZB1 I3M4A5 G1T3N0 W5M6L3 H0XDL1
EC Number
3.1.4.-
Pubmed
19121390
28756777
26354079
22118469
18362917
19820115
+ More
24845553 20566863 17510324 21719571 20798317 17994087 24508170 30249741 17550304 18057021 15632085 23185243 21347285 27289101 29403074 14702039 16710414 15489334 26095358 22722832 16341006 16136131 20431018 22398555 22751099 21993624 19393038 17381049
24845553 20566863 17510324 21719571 20798317 17994087 24508170 30249741 17550304 18057021 15632085 23185243 21347285 27289101 29403074 14702039 16710414 15489334 26095358 22722832 16341006 16136131 20431018 22398555 22751099 21993624 19393038 17381049
EMBL
BABH01023327
KZ150019
PZC74931.1
ODYU01007162
SOQ49700.1
RSAL01000531
+ More
RVE41444.1 KQ461198 KPJ05996.1 KQ459185 KPJ03393.1 AGBW02014553 OWR41446.1 NWSH01002108 PCG69156.1 ODYU01005151 SOQ45727.1 KQ458714 KPJ05473.1 AGBW02012853 OWR44329.1 KQ971380 KYB25011.1 KK852603 KDR20460.1 KQ459930 KPJ19294.1 GGMS01009708 MBY78911.1 DS235853 EEB18824.1 KQ435040 KZC14166.1 CH477439 EAT40899.1 GFXV01008180 MBW19985.1 KQ981864 KYN34228.1 KQ979568 KYN20910.1 KQ976780 KYN08356.1 GL888818 EGI57918.1 OUUW01000012 SPP87455.1 GL440813 EFN65390.1 KQ976662 KYM78577.1 KQ982314 KYQ57884.1 CH916366 EDV96025.1 KK107681 QOIP01000006 EZA48054.1 RLU22212.1 CM000159 EDW93963.2 CH964095 EDW78964.2 KZ288301 PBC28838.1 CH902618 EDV39077.2 KPU77861.1 CH940647 EDW69404.2 CH479240 EDW37540.1 CH379069 EAL29608.3 KRT08182.1 KRF84365.1 KRF84366.1 KRF84367.1 ADTU01014853 ADTU01014854 ADTU01014855 CP012525 ALC44232.1 GECZ01023204 GECZ01017909 JAS46565.1 JAS51860.1 KQ435710 KOX79790.1 UFQS01000091 UFQT01000091 SSW99169.1 SSX19551.1 GBYB01000952 JAG70719.1 LJIJ01000099 ODN02678.1 PYGN01001292 PSN35949.1 GDIP01233722 JAI89679.1 GDIP01186289 LRGB01000966 JAJ37113.1 KZS14351.1 GDIQ01072105 JAN22632.1 GDIQ01021143 JAN73594.1 ACPB03013302 ACPB03013303 GDIP01158300 JAJ65102.1 GDIP01007721 JAM95994.1 GDIQ01195648 JAK56077.1 Y08134 AK316500 AL512288 CH471059 BC014444 AJFE02099117 AJFE02099118 AJFE02099119 AAEX03000411 AACZ04072219 NBAG03000438 PNI24533.1 AK301349 BAG62896.1 KE163683 EPQ13238.1 AAMC01091905 AAMC01091906 CABD030001953 CABD030001954 GABZ01005998 JAA47527.1 JH881135 ELR56305.1 AAPE02033945 AAPE02033946 BC149924 AAI49925.1 LSMT01000400 PFX18637.1 RJVU01053528 ROL33021.1 AGCU01165217 AGCU01165218 GDRN01094955 GDRN01094951 JAI59632.1 ABGA01202045 ABGA01202046 ABGA01202047 ABGA01202048 ABGA01202049 ABGA01365269 ABGA01365270 AGTP01007884 AGTP01007885 AGTP01007886 AAGW02062808 AHAT01009518 AAQR03155980
RVE41444.1 KQ461198 KPJ05996.1 KQ459185 KPJ03393.1 AGBW02014553 OWR41446.1 NWSH01002108 PCG69156.1 ODYU01005151 SOQ45727.1 KQ458714 KPJ05473.1 AGBW02012853 OWR44329.1 KQ971380 KYB25011.1 KK852603 KDR20460.1 KQ459930 KPJ19294.1 GGMS01009708 MBY78911.1 DS235853 EEB18824.1 KQ435040 KZC14166.1 CH477439 EAT40899.1 GFXV01008180 MBW19985.1 KQ981864 KYN34228.1 KQ979568 KYN20910.1 KQ976780 KYN08356.1 GL888818 EGI57918.1 OUUW01000012 SPP87455.1 GL440813 EFN65390.1 KQ976662 KYM78577.1 KQ982314 KYQ57884.1 CH916366 EDV96025.1 KK107681 QOIP01000006 EZA48054.1 RLU22212.1 CM000159 EDW93963.2 CH964095 EDW78964.2 KZ288301 PBC28838.1 CH902618 EDV39077.2 KPU77861.1 CH940647 EDW69404.2 CH479240 EDW37540.1 CH379069 EAL29608.3 KRT08182.1 KRF84365.1 KRF84366.1 KRF84367.1 ADTU01014853 ADTU01014854 ADTU01014855 CP012525 ALC44232.1 GECZ01023204 GECZ01017909 JAS46565.1 JAS51860.1 KQ435710 KOX79790.1 UFQS01000091 UFQT01000091 SSW99169.1 SSX19551.1 GBYB01000952 JAG70719.1 LJIJ01000099 ODN02678.1 PYGN01001292 PSN35949.1 GDIP01233722 JAI89679.1 GDIP01186289 LRGB01000966 JAJ37113.1 KZS14351.1 GDIQ01072105 JAN22632.1 GDIQ01021143 JAN73594.1 ACPB03013302 ACPB03013303 GDIP01158300 JAJ65102.1 GDIP01007721 JAM95994.1 GDIQ01195648 JAK56077.1 Y08134 AK316500 AL512288 CH471059 BC014444 AJFE02099117 AJFE02099118 AJFE02099119 AAEX03000411 AACZ04072219 NBAG03000438 PNI24533.1 AK301349 BAG62896.1 KE163683 EPQ13238.1 AAMC01091905 AAMC01091906 CABD030001953 CABD030001954 GABZ01005998 JAA47527.1 JH881135 ELR56305.1 AAPE02033945 AAPE02033946 BC149924 AAI49925.1 LSMT01000400 PFX18637.1 RJVU01053528 ROL33021.1 AGCU01165217 AGCU01165218 GDRN01094955 GDRN01094951 JAI59632.1 ABGA01202045 ABGA01202046 ABGA01202047 ABGA01202048 ABGA01202049 ABGA01365269 ABGA01365270 AGTP01007884 AGTP01007885 AGTP01007886 AAGW02062808 AHAT01009518 AAQR03155980
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
UP000218220
+ More
UP000007266 UP000027135 UP000192223 UP000009046 UP000076502 UP000008820 UP000078541 UP000078492 UP000078542 UP000007755 UP000268350 UP000000311 UP000078540 UP000075809 UP000001070 UP000053097 UP000279307 UP000002282 UP000007798 UP000242457 UP000005203 UP000007801 UP000008792 UP000008744 UP000001819 UP000005205 UP000092553 UP000053105 UP000094527 UP000245037 UP000076858 UP000015103 UP000007875 UP000005640 UP000240080 UP000002254 UP000265300 UP000261681 UP000002277 UP000189704 UP000265000 UP000008143 UP000001519 UP000286640 UP000001074 UP000009136 UP000225706 UP000264800 UP000007267 UP000265020 UP000001595 UP000248484 UP000189705 UP000005215 UP000001811 UP000018468 UP000005225
UP000007266 UP000027135 UP000192223 UP000009046 UP000076502 UP000008820 UP000078541 UP000078492 UP000078542 UP000007755 UP000268350 UP000000311 UP000078540 UP000075809 UP000001070 UP000053097 UP000279307 UP000002282 UP000007798 UP000242457 UP000005203 UP000007801 UP000008792 UP000008744 UP000001819 UP000005205 UP000092553 UP000053105 UP000094527 UP000245037 UP000076858 UP000015103 UP000007875 UP000005640 UP000240080 UP000002254 UP000265300 UP000261681 UP000002277 UP000189704 UP000265000 UP000008143 UP000001519 UP000286640 UP000001074 UP000009136 UP000225706 UP000264800 UP000007267 UP000265020 UP000001595 UP000248484 UP000189705 UP000005215 UP000001811 UP000018468 UP000005225
Pfam
PF00149 Metallophos
Interpro
Gene 3D
ProteinModelPortal
H9JEU8
A0A2W1BKX4
A0A2H1W9F8
A0A3S2N4T6
A0A194QR38
A0A194QCX4
+ More
A0A212EIY4 A0A2A4JBL4 A0A2H1VY23 A0A194QIT8 A0A212ES85 A0A139WAW8 A0A067RJ29 A0A0N1IQ10 A0A1W4X5I9 A0A2S2QMJ5 E0VZL8 A0A154PQH5 Q172E7 A0A2H8U085 A0A195F186 A0A195E6T8 A0A151IQE7 F4X6F8 A0A3B0JZE6 E2AMG6 A0A195B2Q9 A0A151XBZ8 B4J2N4 A0A026VYW8 B4PDW7 B4N490 A0A2A3EAU8 A0A088AER8 B3MA63 B4LG96 B4HAE2 Q2LZA0 A0A0Q9WI47 A0A158NGE5 A0A0M4F0F5 A0A1B6FNW2 A0A0M9A977 A0A336LRH4 A0A0C9QK20 A0A1D2NBQ4 A0A2P8XVA8 A0A0P4Y4T5 A0A0N8A3F6 A0A0P6DYQ7 A0A0P6HED9 T1HWJ8 A0A0P5DS43 A0A0P6C7J9 A0A0P5JPY0 H2YX51 Q92485 A0A2R9AWT9 J9NYQ3 A0A340XCA0 A0A383Z5Z0 A0A2J8JP43 B4DW34 A0A1U7U7J5 A0A3Q2NN50 S7PQU8 F7C9L5 G3RG15 K9IL01 A0A3Q7TVF7 L8IID3 G1QCT9 A6QQN6 A0A2B4RQL5 A0A3N0Y311 A0A3Q3G0E0 K7GCP6 A0A3Q2EAZ6 A0A0N7ZAS1 H2N8B7 A0A2Y9SVS0 A0A1U8DZB1 I3M4A5 G1T3N0 W5M6L3 H0XDL1
A0A212EIY4 A0A2A4JBL4 A0A2H1VY23 A0A194QIT8 A0A212ES85 A0A139WAW8 A0A067RJ29 A0A0N1IQ10 A0A1W4X5I9 A0A2S2QMJ5 E0VZL8 A0A154PQH5 Q172E7 A0A2H8U085 A0A195F186 A0A195E6T8 A0A151IQE7 F4X6F8 A0A3B0JZE6 E2AMG6 A0A195B2Q9 A0A151XBZ8 B4J2N4 A0A026VYW8 B4PDW7 B4N490 A0A2A3EAU8 A0A088AER8 B3MA63 B4LG96 B4HAE2 Q2LZA0 A0A0Q9WI47 A0A158NGE5 A0A0M4F0F5 A0A1B6FNW2 A0A0M9A977 A0A336LRH4 A0A0C9QK20 A0A1D2NBQ4 A0A2P8XVA8 A0A0P4Y4T5 A0A0N8A3F6 A0A0P6DYQ7 A0A0P6HED9 T1HWJ8 A0A0P5DS43 A0A0P6C7J9 A0A0P5JPY0 H2YX51 Q92485 A0A2R9AWT9 J9NYQ3 A0A340XCA0 A0A383Z5Z0 A0A2J8JP43 B4DW34 A0A1U7U7J5 A0A3Q2NN50 S7PQU8 F7C9L5 G3RG15 K9IL01 A0A3Q7TVF7 L8IID3 G1QCT9 A6QQN6 A0A2B4RQL5 A0A3N0Y311 A0A3Q3G0E0 K7GCP6 A0A3Q2EAZ6 A0A0N7ZAS1 H2N8B7 A0A2Y9SVS0 A0A1U8DZB1 I3M4A5 G1T3N0 W5M6L3 H0XDL1
PDB
5EBE
E-value=7.91003e-49,
Score=490
Ontologies
GO
Topology
Subcellular location
Secreted
Cell membrane
Cell membrane
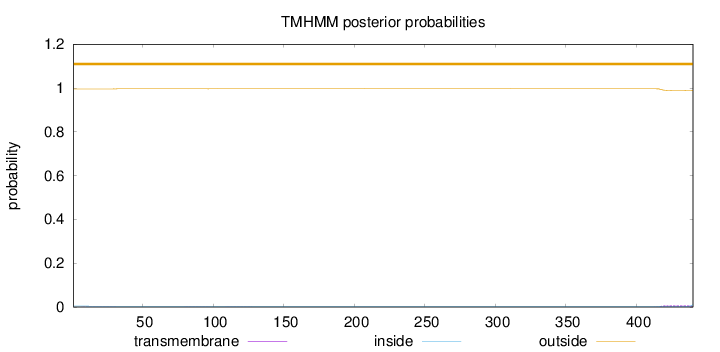
Length:
440
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19457
Exp number, first 60 AAs:
0.01306
Total prob of N-in:
0.00417
outside
1 - 440
Population Genetic Test Statistics
Pi
244.208074
Theta
182.255796
Tajima's D
0.533582
CLR
0.403522
CSRT
0.533523323833808
Interpretation
Uncertain
