Gene
KWMTBOMO04850 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008042
Annotation
PREDICTED:_carbonic_anhydrase_2-like_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.378 Extracellular Reliability : 1.27
Sequence
CDS
ATGTCTCATGTCACGGCGGCCGAGCACATTTCAAGATTAAGACCTTCACAATCACCGATTGCTATATCATTGAGCCGGTGTCCGACTTGGTCTTCGCTCGATCCTCTCAAGTTCAAGGGATATTGGGACAGCAACGCTAACGCCATACTTTTAAATAATGGATCTACTGCATATTTCACGTTCAACGACGCGTCAGTGAGACCGACCCTCAGCGGGGGCCCCCTGATCGGAGAGTACATCTTCGAGCAGATGCACTTCCACTGGTCCGTAGATGACTTCACCGGATGCGAGCACGTGCTCGATGGCCACGGGTACGCCGCAGAATGCCACTTCGTCCACTATAACAGTAAATACGAATCCTTGGAGACGGCGGTGGGCCATCCCGATGGATTGGCTGTGGTCGGATTCCTCCTGGAGACTGTCGACGCTCCCAACCCGAGGTTCGATAGACTAGTACAAGGTCTGGAGGGGATTCAGAAGAGGGAGTCAGTCATGAATGTTACTTCAGAATCCCTATTATGGATGGACAGAGAAGACCTTCAAATCGGCAACTACGTGACTTACAAGGGCTCCCTGACGACACCGCCCTACACCGAGTGTGTCACTTGGATCATTTACGAGAAACCTGTACAAATTGGTTCCGAACAGCTGGGTCTACTTCGACAACTTGAGGGGCCGGACAGTCAACCCATTGAGAGAAACGTTCGTCCGACCCAGAGACACCCGCCGGGCCACTCCGTTATTTACGTTAAGCAAGTTAGGTCAAAACTCTGA
Protein
MSHVTAAEHISRLRPSQSPIAISLSRCPTWSSLDPLKFKGYWDSNANAILLNNGSTAYFTFNDASVRPTLSGGPLIGEYIFEQMHFHWSVDDFTGCEHVLDGHGYAAECHFVHYNSKYESLETAVGHPDGLAVVGFLLETVDAPNPRFDRLVQGLEGIQKRESVMNVTSESLLWMDREDLQIGNYVTYKGSLTTPPYTECVTWIIYEKPVQIGSEQLGLLRQLEGPDSQPIERNVRPTQRHPPGHSVIYVKQVRSKL
Summary
Similarity
Belongs to the alpha-carbonic anhydrase family.
Uniprot
H9JEU5
A0A2W1BT69
A0A2A4JI97
A0A194QKT0
A0A194QDA2
A0A212FNW8
+ More
A0A2H1VHJ7 A0A2A4IW89 A0A2W1BG76 A0A023ELD9 Q179S3 B0WWT4 A0A1Q3FKL8 A0A182Q233 A0A182IX08 D6W9D1 A0A182RUE4 A0A182KEF9 A0A084WJC8 A0A182LB71 Q7Q9S6 A0A182HPC3 A0A182NLY1 Q7PTT6 A0A182VNW5 A0A182P5I1 A0A182YJ04 A0A182M025 A0A182X4C0 A0A1W4XQD7 A0A182WB81 J3JTH8 W5J522 A0A182FGG2 A0A182RKY0 A0A1I8PS75 A0A182PT48 A0A1J1HQJ8 B4JLT6 B4M1A6 A0A0M4ET70 T1PCI3 B4L3A9 A0A3B0K2U7 B3MZD5 A0A182U341 B4GUV5 A0A182W5T0 Q29GS7 B4Q087 A0A034W782 B4MJ47 B4R6K9 B4IJZ5 Q9W3C8 A0A0A1XRF4 W8C1C7 B3NUL6 A0A1W4UDG8 A0A182ITV0 A0A1B0EYC7 T1GZ69 A0A182IAV0 A0A182YCS8 A0A182N0K7 A0A182K827 A0A1B0BMY4 Q7QFC1 A0A1B0F955 A0A0L0CIY0 A0A182SIE2 A0A1L8DFA6 A0A1L8DFB9 A0A182XJW9 A0A182U7Y6 A0A182UW48 A0A2G9P1H5 A0A0K8TMW6 A0A2G9P1E7 K7G9L1 A0A182M1B3 A0A091UNS6 A0A093NS70 A0A091N1G8 A0A084VX56 A0A087QVT7 A0A093FLN1 A0A2I0MWT1 A0A1B0GJC2 A0A1V4KGV8 A0A093EIF1 A0A060XWT2 A0A091KYH0 A0A091TY22 U3I3N7 A0A091FY17 R0KEY7 H1A5U6 A0A091PFP3
A0A2H1VHJ7 A0A2A4IW89 A0A2W1BG76 A0A023ELD9 Q179S3 B0WWT4 A0A1Q3FKL8 A0A182Q233 A0A182IX08 D6W9D1 A0A182RUE4 A0A182KEF9 A0A084WJC8 A0A182LB71 Q7Q9S6 A0A182HPC3 A0A182NLY1 Q7PTT6 A0A182VNW5 A0A182P5I1 A0A182YJ04 A0A182M025 A0A182X4C0 A0A1W4XQD7 A0A182WB81 J3JTH8 W5J522 A0A182FGG2 A0A182RKY0 A0A1I8PS75 A0A182PT48 A0A1J1HQJ8 B4JLT6 B4M1A6 A0A0M4ET70 T1PCI3 B4L3A9 A0A3B0K2U7 B3MZD5 A0A182U341 B4GUV5 A0A182W5T0 Q29GS7 B4Q087 A0A034W782 B4MJ47 B4R6K9 B4IJZ5 Q9W3C8 A0A0A1XRF4 W8C1C7 B3NUL6 A0A1W4UDG8 A0A182ITV0 A0A1B0EYC7 T1GZ69 A0A182IAV0 A0A182YCS8 A0A182N0K7 A0A182K827 A0A1B0BMY4 Q7QFC1 A0A1B0F955 A0A0L0CIY0 A0A182SIE2 A0A1L8DFA6 A0A1L8DFB9 A0A182XJW9 A0A182U7Y6 A0A182UW48 A0A2G9P1H5 A0A0K8TMW6 A0A2G9P1E7 K7G9L1 A0A182M1B3 A0A091UNS6 A0A093NS70 A0A091N1G8 A0A084VX56 A0A087QVT7 A0A093FLN1 A0A2I0MWT1 A0A1B0GJC2 A0A1V4KGV8 A0A093EIF1 A0A060XWT2 A0A091KYH0 A0A091TY22 U3I3N7 A0A091FY17 R0KEY7 H1A5U6 A0A091PFP3
Pubmed
19121390
28756777
26354079
22118469
24945155
26483478
+ More
17510324 18362917 19820115 24438588 20966253 12364791 14747013 17210077 9087549 25244985 22516182 23537049 20920257 23761445 17994087 18057021 25315136 15632085 23185243 17550304 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 24495485 26108605 26369729 17381049 23371554 24755649 23749191 20360741
17510324 18362917 19820115 24438588 20966253 12364791 14747013 17210077 9087549 25244985 22516182 23537049 20920257 23761445 17994087 18057021 25315136 15632085 23185243 17550304 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 24495485 26108605 26369729 17381049 23371554 24755649 23749191 20360741
EMBL
BABH01023309
BABH01023310
BABH01023311
BABH01023312
KZ150019
PZC74923.1
+ More
NWSH01001366 PCG71519.1 KQ461198 KPJ06004.1 KQ459185 KPJ03399.1 AGBW02004596 OWR55390.1 ODYU01002356 SOQ39744.1 NWSH01005409 PCG64237.1 KZ150067 PZC74129.1 JXUM01029962 JXUM01029963 JXUM01029964 JXUM01029965 GAPW01003562 KQ560869 JAC10036.1 KXJ80553.1 CH477346 EAT42998.1 EJY57590.1 DS232152 EDS36195.1 GFDL01006916 JAV28129.1 AXCN02000954 KQ971312 EEZ98179.1 ATLV01024013 KE525348 KFB50322.1 AY280613 AAAB01008900 AAQ21366.1 EAA09468.3 APCN01002969 AAAB01008400 EAA03371.4 AXCM01005738 APGK01030408 APGK01030409 BT126535 KB740771 KB632375 AEE61499.1 ENN78981.1 ERL93862.1 ADMH02002104 ETN59066.1 CVRI01000010 CRK88750.1 CH916371 EDV91697.1 CH940651 EDW65460.1 KRF82241.1 CP012528 ALC48701.1 KA645850 AFP60479.1 CH933810 EDW07037.1 OUUW01000015 SPP88605.1 CH902635 EDV33736.2 KPU74729.1 KPU74730.1 CH479192 EDW26492.1 CH379064 EAL32032.1 KRT06788.1 CM000162 EDX02224.1 GAKP01008952 JAC50000.1 CH963738 EDW72136.1 CM000366 EDX17439.1 CH480850 EDW51351.1 AE014298 AY069607 AAF46402.1 AAL39752.1 AGB95192.1 GBXI01013509 GBXI01000691 JAD00783.1 JAD13601.1 GAMC01007020 GAMC01007019 JAB99536.1 CH954180 EDV46339.1 AJVK01033737 AJVK01033738 CAQQ02161122 CAQQ02161123 APCN01001344 JXJN01017114 AAAB01008846 EAA06713.4 CCAG010018936 JRES01000336 KNC32177.1 GFDF01008942 JAV05142.1 GFDF01008943 JAV05141.1 KV922996 PIN97164.1 GDAI01001914 JAI15689.1 PIN97165.1 AGCU01018659 AGCU01018660 AXCM01000769 KL409868 KFQ92609.1 KL224729 KFW62997.1 KK840822 KFP83321.1 ATLV01017849 KE525195 KFB42550.1 KL225953 KFM05341.1 KK634529 KFV57570.1 AKCR02000001 PKK34127.1 AJWK01020126 AJWK01020127 LSYS01003169 OPJ83621.1 KL462937 KFV14321.1 FR905752 CDQ81390.1 KK759294 KFP44340.1 KK467158 KFQ82490.1 ADON01001530 ADON01001531 ADON01052734 ADON01052735 KL447477 KFO74084.1 KB742403 EOB08991.1 ABQF01005621 KK673224 KFQ06722.1
NWSH01001366 PCG71519.1 KQ461198 KPJ06004.1 KQ459185 KPJ03399.1 AGBW02004596 OWR55390.1 ODYU01002356 SOQ39744.1 NWSH01005409 PCG64237.1 KZ150067 PZC74129.1 JXUM01029962 JXUM01029963 JXUM01029964 JXUM01029965 GAPW01003562 KQ560869 JAC10036.1 KXJ80553.1 CH477346 EAT42998.1 EJY57590.1 DS232152 EDS36195.1 GFDL01006916 JAV28129.1 AXCN02000954 KQ971312 EEZ98179.1 ATLV01024013 KE525348 KFB50322.1 AY280613 AAAB01008900 AAQ21366.1 EAA09468.3 APCN01002969 AAAB01008400 EAA03371.4 AXCM01005738 APGK01030408 APGK01030409 BT126535 KB740771 KB632375 AEE61499.1 ENN78981.1 ERL93862.1 ADMH02002104 ETN59066.1 CVRI01000010 CRK88750.1 CH916371 EDV91697.1 CH940651 EDW65460.1 KRF82241.1 CP012528 ALC48701.1 KA645850 AFP60479.1 CH933810 EDW07037.1 OUUW01000015 SPP88605.1 CH902635 EDV33736.2 KPU74729.1 KPU74730.1 CH479192 EDW26492.1 CH379064 EAL32032.1 KRT06788.1 CM000162 EDX02224.1 GAKP01008952 JAC50000.1 CH963738 EDW72136.1 CM000366 EDX17439.1 CH480850 EDW51351.1 AE014298 AY069607 AAF46402.1 AAL39752.1 AGB95192.1 GBXI01013509 GBXI01000691 JAD00783.1 JAD13601.1 GAMC01007020 GAMC01007019 JAB99536.1 CH954180 EDV46339.1 AJVK01033737 AJVK01033738 CAQQ02161122 CAQQ02161123 APCN01001344 JXJN01017114 AAAB01008846 EAA06713.4 CCAG010018936 JRES01000336 KNC32177.1 GFDF01008942 JAV05142.1 GFDF01008943 JAV05141.1 KV922996 PIN97164.1 GDAI01001914 JAI15689.1 PIN97165.1 AGCU01018659 AGCU01018660 AXCM01000769 KL409868 KFQ92609.1 KL224729 KFW62997.1 KK840822 KFP83321.1 ATLV01017849 KE525195 KFB42550.1 KL225953 KFM05341.1 KK634529 KFV57570.1 AKCR02000001 PKK34127.1 AJWK01020126 AJWK01020127 LSYS01003169 OPJ83621.1 KL462937 KFV14321.1 FR905752 CDQ81390.1 KK759294 KFP44340.1 KK467158 KFQ82490.1 ADON01001530 ADON01001531 ADON01052734 ADON01052735 KL447477 KFO74084.1 KB742403 EOB08991.1 ABQF01005621 KK673224 KFQ06722.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000069940
+ More
UP000249989 UP000008820 UP000002320 UP000075886 UP000075880 UP000007266 UP000075900 UP000075881 UP000030765 UP000075882 UP000007062 UP000075840 UP000075884 UP000075903 UP000075885 UP000076408 UP000075883 UP000076407 UP000192223 UP000075920 UP000019118 UP000030742 UP000000673 UP000069272 UP000095300 UP000183832 UP000001070 UP000008792 UP000092553 UP000095301 UP000009192 UP000268350 UP000007801 UP000075902 UP000008744 UP000001819 UP000002282 UP000007798 UP000000304 UP000001292 UP000000803 UP000008711 UP000192221 UP000092462 UP000015102 UP000092460 UP000092444 UP000037069 UP000075901 UP000007267 UP000053283 UP000054081 UP000053286 UP000053872 UP000092461 UP000190648 UP000193380 UP000016666 UP000053760 UP000007754
UP000249989 UP000008820 UP000002320 UP000075886 UP000075880 UP000007266 UP000075900 UP000075881 UP000030765 UP000075882 UP000007062 UP000075840 UP000075884 UP000075903 UP000075885 UP000076408 UP000075883 UP000076407 UP000192223 UP000075920 UP000019118 UP000030742 UP000000673 UP000069272 UP000095300 UP000183832 UP000001070 UP000008792 UP000092553 UP000095301 UP000009192 UP000268350 UP000007801 UP000075902 UP000008744 UP000001819 UP000002282 UP000007798 UP000000304 UP000001292 UP000000803 UP000008711 UP000192221 UP000092462 UP000015102 UP000092460 UP000092444 UP000037069 UP000075901 UP000007267 UP000053283 UP000054081 UP000053286 UP000053872 UP000092461 UP000190648 UP000193380 UP000016666 UP000053760 UP000007754
Pfam
PF00194 Carb_anhydrase
Interpro
SUPFAM
SSF51069
SSF51069
Gene 3D
ProteinModelPortal
H9JEU5
A0A2W1BT69
A0A2A4JI97
A0A194QKT0
A0A194QDA2
A0A212FNW8
+ More
A0A2H1VHJ7 A0A2A4IW89 A0A2W1BG76 A0A023ELD9 Q179S3 B0WWT4 A0A1Q3FKL8 A0A182Q233 A0A182IX08 D6W9D1 A0A182RUE4 A0A182KEF9 A0A084WJC8 A0A182LB71 Q7Q9S6 A0A182HPC3 A0A182NLY1 Q7PTT6 A0A182VNW5 A0A182P5I1 A0A182YJ04 A0A182M025 A0A182X4C0 A0A1W4XQD7 A0A182WB81 J3JTH8 W5J522 A0A182FGG2 A0A182RKY0 A0A1I8PS75 A0A182PT48 A0A1J1HQJ8 B4JLT6 B4M1A6 A0A0M4ET70 T1PCI3 B4L3A9 A0A3B0K2U7 B3MZD5 A0A182U341 B4GUV5 A0A182W5T0 Q29GS7 B4Q087 A0A034W782 B4MJ47 B4R6K9 B4IJZ5 Q9W3C8 A0A0A1XRF4 W8C1C7 B3NUL6 A0A1W4UDG8 A0A182ITV0 A0A1B0EYC7 T1GZ69 A0A182IAV0 A0A182YCS8 A0A182N0K7 A0A182K827 A0A1B0BMY4 Q7QFC1 A0A1B0F955 A0A0L0CIY0 A0A182SIE2 A0A1L8DFA6 A0A1L8DFB9 A0A182XJW9 A0A182U7Y6 A0A182UW48 A0A2G9P1H5 A0A0K8TMW6 A0A2G9P1E7 K7G9L1 A0A182M1B3 A0A091UNS6 A0A093NS70 A0A091N1G8 A0A084VX56 A0A087QVT7 A0A093FLN1 A0A2I0MWT1 A0A1B0GJC2 A0A1V4KGV8 A0A093EIF1 A0A060XWT2 A0A091KYH0 A0A091TY22 U3I3N7 A0A091FY17 R0KEY7 H1A5U6 A0A091PFP3
A0A2H1VHJ7 A0A2A4IW89 A0A2W1BG76 A0A023ELD9 Q179S3 B0WWT4 A0A1Q3FKL8 A0A182Q233 A0A182IX08 D6W9D1 A0A182RUE4 A0A182KEF9 A0A084WJC8 A0A182LB71 Q7Q9S6 A0A182HPC3 A0A182NLY1 Q7PTT6 A0A182VNW5 A0A182P5I1 A0A182YJ04 A0A182M025 A0A182X4C0 A0A1W4XQD7 A0A182WB81 J3JTH8 W5J522 A0A182FGG2 A0A182RKY0 A0A1I8PS75 A0A182PT48 A0A1J1HQJ8 B4JLT6 B4M1A6 A0A0M4ET70 T1PCI3 B4L3A9 A0A3B0K2U7 B3MZD5 A0A182U341 B4GUV5 A0A182W5T0 Q29GS7 B4Q087 A0A034W782 B4MJ47 B4R6K9 B4IJZ5 Q9W3C8 A0A0A1XRF4 W8C1C7 B3NUL6 A0A1W4UDG8 A0A182ITV0 A0A1B0EYC7 T1GZ69 A0A182IAV0 A0A182YCS8 A0A182N0K7 A0A182K827 A0A1B0BMY4 Q7QFC1 A0A1B0F955 A0A0L0CIY0 A0A182SIE2 A0A1L8DFA6 A0A1L8DFB9 A0A182XJW9 A0A182U7Y6 A0A182UW48 A0A2G9P1H5 A0A0K8TMW6 A0A2G9P1E7 K7G9L1 A0A182M1B3 A0A091UNS6 A0A093NS70 A0A091N1G8 A0A084VX56 A0A087QVT7 A0A093FLN1 A0A2I0MWT1 A0A1B0GJC2 A0A1V4KGV8 A0A093EIF1 A0A060XWT2 A0A091KYH0 A0A091TY22 U3I3N7 A0A091FY17 R0KEY7 H1A5U6 A0A091PFP3
PDB
6B00
E-value=5.31045e-38,
Score=393
Ontologies
PATHWAY
PANTHER
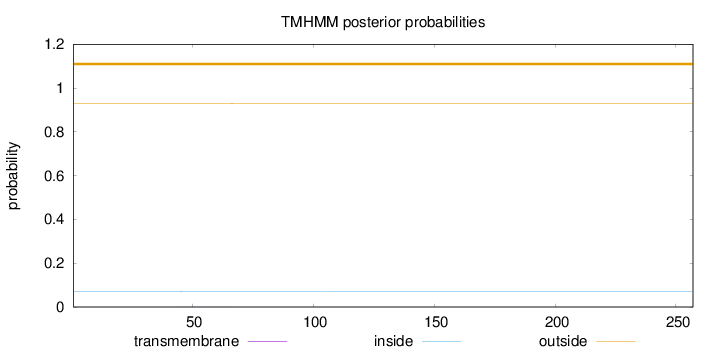
Topology
Length:
257
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0116
Exp number, first 60 AAs:
0.00725
Total prob of N-in:
0.07013
outside
1 - 257
Population Genetic Test Statistics
Pi
306.126664
Theta
180.373048
Tajima's D
1.826278
CLR
0.616169
CSRT
0.852057397130144
Interpretation
Uncertain
