Gene
KWMTBOMO04835 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013945
Annotation
actin?_muscle-type_A2_[Bombyx_mori]
Full name
Actin, muscle-type A2
+ More
Actin, indirect flight muscle
Actin, indirect flight muscle
Alternative Name
Actin-88F
Location in the cell
Cytoplasmic Reliability : 3.817
Sequence
CDS
ATGTGTGACGATGATGCGGGCGCGCTCGTCGTCGACAACGGATCCGGCATGTGCAAGGCTGGATTCGCGGGGGACGACGCCCCTCGAGCCGTGTTCCCTTCCATAGTGGGCCGCCCGCGCCACCAGGGCGTCATGGTCGGCATGGGCCAAAAAGATTCGTATGTAGGTGACGAAGCCCAGAGCAAGAGAGGTATCCTCACTCTAAAGTACCCCATCGAGCACGGGATAATCACCAACTGGGATGACATGGAGAAGATCTGGCATCACACGTTCTACAATGAGCTGCGCGTCGCTCCCGAAGAGCACCCAATCCTGCTGACCGAGGCGCCCCTGAACCCGAAGGCCAACAGAGAGAAGATGACTCAGATCATGTTCGAGACGTTCAATTGTCCCGCCATGTACGTCGCCATCCAGGCCGTGCTCTCGCTCTACGCCTCCGGTCGTACCACCGGCATCGTGCTCGACTCCGGAGACGGTGTCTCCCACACCGTACCCATCTACGAAGGCTATGCGCTGCCCCACGCCATTCTCCGTCTGGACTTGGCCGGACGCGACTTGACTGACTACCTCATGAAGATCCTCACCGAGAGGGGCTACTCGTTCACTACCACTGCCGAGAGGGAAATCGTCCGCGACATCAAGGAGAAGCTGTGTTATGTCGCTCTCGACTTTGAGCAGGAAATGCAGACTGCGGCCGCTTCAACCTCTCTGGAGAAATCCTACGAGCTACCAGACGGACAGGTGATCACCATCGGCAACGAGCGCTTCCGGTGTCCCGAGGCTCTGTTCCAACCCTCGTTCCTGGGTATGGAGTCGTGCGGCATCCACGAGACCGTGTACAACTCGATTATGAAGTGCGACGTGGACATCCGCAAGGACCTGTACGCCAACACGGTGCTGTCGGGCGGCACGACCATGTACCCCGGTATCGCCGACAGGATGCAGAAGGAGATCACCGCCCTCGCGCCCTCGACCATCAAGATCAAGATCATCGCTCCCCCCGAGAGGAAGTATTCCGTGTGGATCGGCGGCTCGATCCTGGCGTCGCTGTCCACCTTCCAGCAGATGTGGATCTCGAAGCAGGAGTACGACGAGTCCGGGCCGGGCATCGTTCACCGCAAGTGCTTCTAA
Protein
MCDDDAGALVVDNGSGMCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYVGDEAQSKRGILTLKYPIEHGIITNWDDMEKIWHHTFYNELRVAPEEHPILLTEAPLNPKANREKMTQIMFETFNCPAMYVAIQAVLSLYASGRTTGIVLDSGDGVSHTVPIYEGYALPHAILRLDLAGRDLTDYLMKILTERGYSFTTTAEREIVRDIKEKLCYVALDFEQEMQTAAASTSLEKSYELPDGQVITIGNERFRCPEALFQPSFLGMESCGIHETVYNSIMKCDVDIRKDLYANTVLSGGTTMYPGIADRMQKEITALAPSTIKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQEYDESGPGIVHRKCF
Summary
Miscellaneous
There are at least 5 different actin genes in this organism.
In Drosophila there are 6 closely related actin genes.
Abnormalities in Act88F-ifm(3)7 flight muscles result from incorporation of the mutant actin isoform into assembling myofibrils.
In Drosophila there are 6 closely related actin genes.
Abnormalities in Act88F-ifm(3)7 flight muscles result from incorporation of the mutant actin isoform into assembling myofibrils.
Similarity
Belongs to the actin family.
Keywords
Acetylation
ATP-binding
Complete proteome
Cytoplasm
Cytoskeleton
Nucleotide-binding
Oxidation
Reference proteome
Methylation
Muscle protein
Feature
propeptide Removed in mature form
chain Actin, muscle-type A2
chain Actin, muscle-type A2
Uniprot
A0A2W1BJV8
A0A2A4J2T2
A0A2H1WBG9
P07837
F5BYH6
Q9NA03
+ More
E9FRT1 A0A0L7KTD0 A0A1J1HPZ0 A0A0P5F1P7 A0A0P5C5X9 A0A0N8A3C7 A0A0N8AGL2 F2D4P0 A0A1W4VNM5 A0A224XRR1 Q5XXT4 A0A1B0G562 Q3HNA6 A0A125RA11 A0A2D1QW10 A0A2H8TVL9 A0A1A9XG84 D3KU63 A0A1S3CVP9 A0A3G4R8Q5 A0A3G1DKW0 A0A3B0JL71 A0A0P4VVE9 A0A1A9VBA9 A0A1B0BNN7 B5DVZ6 A0A140EFS7 B6IDW7 Q5XUA4 B4PRP2 A0A0V0G386 X1X262 A0A170ZGA9 Q6PPI5 A0A2J7RHY5 A1BPP6 A0A067QL86 M9VDW4 A7WPD3 A0A1S5RIQ2 A0A1I8Q8X9 B3LZT0 B4N9R8 A0A0L0CD38 A0A125RA09 A0A2Z5RE76 B3P3R7 A0A0N8CDK3 A0A1A9ZZJ7 A0A1A9WJB9 A0A1Y1LUA6 R4FQA8 C9VXR9 A0A0P5CHV2 A0A1B6EXE0 T1PDA4 E0VKP4 A0A1B6CK57 A0A0A9W354 P83968 P83967 P83969 A0A1L8E4X1 A0A1B0D7E8 A0A151JMQ0 A0A1W4XH28 A0A1W4XWB9 A0A2Z4G633 A0A151I4W0 K7J803 X5CT98 A0A195FX88 A0A2R4GMB1 A0A158NXU2 A0A0P5CGV3 A0A3G1SUZ3 A0A151WIN4 A0A0P5CHD6 A0A0N8A945 A0A1L8E5H6 Q7Q7K6 A0A0P5EGX5 A0A2M4ATG0 A0A0H4IV48 Q5RLJ4 V5GR51 A0A023J916 D6WF19 A0A023VZM4 C8CD00 A0A0P5DTY5 A0A026X3P0 N6TH73 A0A1B1UZE0
E9FRT1 A0A0L7KTD0 A0A1J1HPZ0 A0A0P5F1P7 A0A0P5C5X9 A0A0N8A3C7 A0A0N8AGL2 F2D4P0 A0A1W4VNM5 A0A224XRR1 Q5XXT4 A0A1B0G562 Q3HNA6 A0A125RA11 A0A2D1QW10 A0A2H8TVL9 A0A1A9XG84 D3KU63 A0A1S3CVP9 A0A3G4R8Q5 A0A3G1DKW0 A0A3B0JL71 A0A0P4VVE9 A0A1A9VBA9 A0A1B0BNN7 B5DVZ6 A0A140EFS7 B6IDW7 Q5XUA4 B4PRP2 A0A0V0G386 X1X262 A0A170ZGA9 Q6PPI5 A0A2J7RHY5 A1BPP6 A0A067QL86 M9VDW4 A7WPD3 A0A1S5RIQ2 A0A1I8Q8X9 B3LZT0 B4N9R8 A0A0L0CD38 A0A125RA09 A0A2Z5RE76 B3P3R7 A0A0N8CDK3 A0A1A9ZZJ7 A0A1A9WJB9 A0A1Y1LUA6 R4FQA8 C9VXR9 A0A0P5CHV2 A0A1B6EXE0 T1PDA4 E0VKP4 A0A1B6CK57 A0A0A9W354 P83968 P83967 P83969 A0A1L8E4X1 A0A1B0D7E8 A0A151JMQ0 A0A1W4XH28 A0A1W4XWB9 A0A2Z4G633 A0A151I4W0 K7J803 X5CT98 A0A195FX88 A0A2R4GMB1 A0A158NXU2 A0A0P5CGV3 A0A3G1SUZ3 A0A151WIN4 A0A0P5CHD6 A0A0N8A945 A0A1L8E5H6 Q7Q7K6 A0A0P5EGX5 A0A2M4ATG0 A0A0H4IV48 Q5RLJ4 V5GR51 A0A023J916 D6WF19 A0A023VZM4 C8CD00 A0A0P5DTY5 A0A026X3P0 N6TH73 A0A1B1UZE0
Pubmed
28756777
3562238
21452889
21292972
26227816
21415278
+ More
16537425 26826390 20022415 27483241 27129103 15632085 17994087 26873291 17550304 17469070 24845553 27494615 26108605 26760975 28004739 25315136 20566863 25401762 1511890 6405041 10731132 12537572 6263481 6488317 17353360 22116028 7951267 29889877 20075255 29590372 21347285 12364791 18362917 19820115 24508170 23537049
16537425 26826390 20022415 27483241 27129103 15632085 17994087 26873291 17550304 17469070 24845553 27494615 26108605 26760975 28004739 25315136 20566863 25401762 1511890 6405041 10731132 12537572 6263481 6488317 17353360 22116028 7951267 29889877 20075255 29590372 21347285 12364791 18362917 19820115 24508170 23537049
EMBL
KZ150126
PZC73116.1
NWSH01003858
PCG65750.1
ODYU01007534
SOQ50387.1
+ More
X06363 JF417981 AEB26312.1 AJ292554 GDIP01175048 GDIP01175045 GDIQ01130636 GDIP01129717 GDIP01037836 GDIQ01037496 GDIQ01037495 CAB99474.1 JAJ48354.1 JAL21090.1 GL732524 GL732523 EFX89657.1 EFX89901.1 JTDY01006084 KOB66311.1 CVRI01000015 CRK90035.1 GDIQ01268830 JAJ82894.1 GDIP01175046 JAJ48356.1 GDIP01186521 JAJ36881.1 GDIP01149442 GDIP01149441 JAJ73961.1 AK358850 BAJ90061.1 GFTR01005593 JAW10833.1 AY725778 AAU95191.1 CCAG010013799 DQ206832 ABA62321.1 KU196670 AMD16550.1 KY038869 ATP16752.1 GFXV01006509 MBW18314.1 AB509262 BAI77996.1 MG993326 AYU58903.1 KU188517 ANE06405.1 OUUW01000007 SPP82975.1 GDKW01000574 JAI56021.1 JXJN01017519 CM000070 EDY67713.1 KT369806 AML25535.1 BT050557 ACJ13264.1 AY737550 AAU84943.1 CM000160 EDW97442.1 GECL01003587 JAP02537.1 ABLF02032889 GEMB01002235 JAS00945.1 AY588061 AAT01073.1 NEVH01003508 PNF40452.1 DQ450899 ABE27979.1 KK853207 KDR09838.1 KC161211 AGJ76539.1 AM850110 CAO98769.1 KX108734 ANV22067.1 CH902617 EDV43074.1 CH964232 EDW80633.1 JRES01000563 KNC30170.1 KU196668 AMD16548.1 FX983744 BAX07243.1 CH954181 EDV49023.1 GDIP01142223 JAL61491.1 GEZM01048387 GEZM01048386 JAV76591.1 ACPB03021529 GAHY01000509 JAA77001.1 EU179846 EU179847 EU179848 ABY48093.1 ABY48095.1 GDIP01175047 GDIP01170376 JAJ53026.1 GECZ01027298 GECZ01011797 JAS42471.1 JAS57972.1 KA646663 KA646664 AFP61293.1 DS235250 EEB13950.1 GEDC01023461 GEDC01016965 GEDC01002583 JAS13837.1 JAS20333.1 JAS34715.1 GBHO01044314 GBRD01005390 JAF99289.1 JAG60431.1 M87274 CM000364 M18826 K02065 AB003910 AE014297 M18830 L12253 GFDF01000313 JAV13771.1 AJVK01003905 KQ978931 KYN27466.1 MG983770 AWV96605.1 KQ976462 KYM84633.1 AAZX01005009 KJ192330 AHW40461.1 KQ981204 KYN44932.1 MG264369 AVT69429.1 ADTU01003540 ADTU01003541 GDIP01170424 JAJ52978.1 MG243593 AXU38338.1 KQ983089 KYQ47686.1 GDIP01175044 GDIP01170375 JAJ53027.1 GDIP01170377 JAJ53025.1 GFDF01000312 JAV13772.1 AAAB01008956 EAA10668.4 GDIP01143275 JAJ80127.1 GGFK01010774 MBW44095.1 KP704257 AKO62881.1 AY817141 AAV65298.1 GALX01004399 JAB64067.1 KF792064 AHH25009.1 KQ971319 EFA00468.1 KJ507199 AHY22050.1 GQ389791 ACV32627.1 GDIP01150736 JAJ72666.1 KK107013 EZA62930.1 APGK01027120 KB740648 KB632292 ENN79784.1 ERL91608.1 KU521368 KX428475 ANS71248.1 ANW09586.1
X06363 JF417981 AEB26312.1 AJ292554 GDIP01175048 GDIP01175045 GDIQ01130636 GDIP01129717 GDIP01037836 GDIQ01037496 GDIQ01037495 CAB99474.1 JAJ48354.1 JAL21090.1 GL732524 GL732523 EFX89657.1 EFX89901.1 JTDY01006084 KOB66311.1 CVRI01000015 CRK90035.1 GDIQ01268830 JAJ82894.1 GDIP01175046 JAJ48356.1 GDIP01186521 JAJ36881.1 GDIP01149442 GDIP01149441 JAJ73961.1 AK358850 BAJ90061.1 GFTR01005593 JAW10833.1 AY725778 AAU95191.1 CCAG010013799 DQ206832 ABA62321.1 KU196670 AMD16550.1 KY038869 ATP16752.1 GFXV01006509 MBW18314.1 AB509262 BAI77996.1 MG993326 AYU58903.1 KU188517 ANE06405.1 OUUW01000007 SPP82975.1 GDKW01000574 JAI56021.1 JXJN01017519 CM000070 EDY67713.1 KT369806 AML25535.1 BT050557 ACJ13264.1 AY737550 AAU84943.1 CM000160 EDW97442.1 GECL01003587 JAP02537.1 ABLF02032889 GEMB01002235 JAS00945.1 AY588061 AAT01073.1 NEVH01003508 PNF40452.1 DQ450899 ABE27979.1 KK853207 KDR09838.1 KC161211 AGJ76539.1 AM850110 CAO98769.1 KX108734 ANV22067.1 CH902617 EDV43074.1 CH964232 EDW80633.1 JRES01000563 KNC30170.1 KU196668 AMD16548.1 FX983744 BAX07243.1 CH954181 EDV49023.1 GDIP01142223 JAL61491.1 GEZM01048387 GEZM01048386 JAV76591.1 ACPB03021529 GAHY01000509 JAA77001.1 EU179846 EU179847 EU179848 ABY48093.1 ABY48095.1 GDIP01175047 GDIP01170376 JAJ53026.1 GECZ01027298 GECZ01011797 JAS42471.1 JAS57972.1 KA646663 KA646664 AFP61293.1 DS235250 EEB13950.1 GEDC01023461 GEDC01016965 GEDC01002583 JAS13837.1 JAS20333.1 JAS34715.1 GBHO01044314 GBRD01005390 JAF99289.1 JAG60431.1 M87274 CM000364 M18826 K02065 AB003910 AE014297 M18830 L12253 GFDF01000313 JAV13771.1 AJVK01003905 KQ978931 KYN27466.1 MG983770 AWV96605.1 KQ976462 KYM84633.1 AAZX01005009 KJ192330 AHW40461.1 KQ981204 KYN44932.1 MG264369 AVT69429.1 ADTU01003540 ADTU01003541 GDIP01170424 JAJ52978.1 MG243593 AXU38338.1 KQ983089 KYQ47686.1 GDIP01175044 GDIP01170375 JAJ53027.1 GDIP01170377 JAJ53025.1 GFDF01000312 JAV13772.1 AAAB01008956 EAA10668.4 GDIP01143275 JAJ80127.1 GGFK01010774 MBW44095.1 KP704257 AKO62881.1 AY817141 AAV65298.1 GALX01004399 JAB64067.1 KF792064 AHH25009.1 KQ971319 EFA00468.1 KJ507199 AHY22050.1 GQ389791 ACV32627.1 GDIP01150736 JAJ72666.1 KK107013 EZA62930.1 APGK01027120 KB740648 KB632292 ENN79784.1 ERL91608.1 KU521368 KX428475 ANS71248.1 ANW09586.1
Proteomes
UP000218220
UP000005204
UP000000305
UP000037510
UP000183832
UP000192221
+ More
UP000092444 UP000092443 UP000079169 UP000268350 UP000078200 UP000092460 UP000001819 UP000002282 UP000007819 UP000235965 UP000027135 UP000095300 UP000007801 UP000007798 UP000037069 UP000008711 UP000092445 UP000091820 UP000015103 UP000095301 UP000009046 UP000000304 UP000000803 UP000092462 UP000078492 UP000192223 UP000078540 UP000002358 UP000078541 UP000005205 UP000075809 UP000007062 UP000007266 UP000053097 UP000019118 UP000030742
UP000092444 UP000092443 UP000079169 UP000268350 UP000078200 UP000092460 UP000001819 UP000002282 UP000007819 UP000235965 UP000027135 UP000095300 UP000007801 UP000007798 UP000037069 UP000008711 UP000092445 UP000091820 UP000015103 UP000095301 UP000009046 UP000000304 UP000000803 UP000092462 UP000078492 UP000192223 UP000078540 UP000002358 UP000078541 UP000005205 UP000075809 UP000007062 UP000007266 UP000053097 UP000019118 UP000030742
Pfam
PF00022 Actin
ProteinModelPortal
A0A2W1BJV8
A0A2A4J2T2
A0A2H1WBG9
P07837
F5BYH6
Q9NA03
+ More
E9FRT1 A0A0L7KTD0 A0A1J1HPZ0 A0A0P5F1P7 A0A0P5C5X9 A0A0N8A3C7 A0A0N8AGL2 F2D4P0 A0A1W4VNM5 A0A224XRR1 Q5XXT4 A0A1B0G562 Q3HNA6 A0A125RA11 A0A2D1QW10 A0A2H8TVL9 A0A1A9XG84 D3KU63 A0A1S3CVP9 A0A3G4R8Q5 A0A3G1DKW0 A0A3B0JL71 A0A0P4VVE9 A0A1A9VBA9 A0A1B0BNN7 B5DVZ6 A0A140EFS7 B6IDW7 Q5XUA4 B4PRP2 A0A0V0G386 X1X262 A0A170ZGA9 Q6PPI5 A0A2J7RHY5 A1BPP6 A0A067QL86 M9VDW4 A7WPD3 A0A1S5RIQ2 A0A1I8Q8X9 B3LZT0 B4N9R8 A0A0L0CD38 A0A125RA09 A0A2Z5RE76 B3P3R7 A0A0N8CDK3 A0A1A9ZZJ7 A0A1A9WJB9 A0A1Y1LUA6 R4FQA8 C9VXR9 A0A0P5CHV2 A0A1B6EXE0 T1PDA4 E0VKP4 A0A1B6CK57 A0A0A9W354 P83968 P83967 P83969 A0A1L8E4X1 A0A1B0D7E8 A0A151JMQ0 A0A1W4XH28 A0A1W4XWB9 A0A2Z4G633 A0A151I4W0 K7J803 X5CT98 A0A195FX88 A0A2R4GMB1 A0A158NXU2 A0A0P5CGV3 A0A3G1SUZ3 A0A151WIN4 A0A0P5CHD6 A0A0N8A945 A0A1L8E5H6 Q7Q7K6 A0A0P5EGX5 A0A2M4ATG0 A0A0H4IV48 Q5RLJ4 V5GR51 A0A023J916 D6WF19 A0A023VZM4 C8CD00 A0A0P5DTY5 A0A026X3P0 N6TH73 A0A1B1UZE0
E9FRT1 A0A0L7KTD0 A0A1J1HPZ0 A0A0P5F1P7 A0A0P5C5X9 A0A0N8A3C7 A0A0N8AGL2 F2D4P0 A0A1W4VNM5 A0A224XRR1 Q5XXT4 A0A1B0G562 Q3HNA6 A0A125RA11 A0A2D1QW10 A0A2H8TVL9 A0A1A9XG84 D3KU63 A0A1S3CVP9 A0A3G4R8Q5 A0A3G1DKW0 A0A3B0JL71 A0A0P4VVE9 A0A1A9VBA9 A0A1B0BNN7 B5DVZ6 A0A140EFS7 B6IDW7 Q5XUA4 B4PRP2 A0A0V0G386 X1X262 A0A170ZGA9 Q6PPI5 A0A2J7RHY5 A1BPP6 A0A067QL86 M9VDW4 A7WPD3 A0A1S5RIQ2 A0A1I8Q8X9 B3LZT0 B4N9R8 A0A0L0CD38 A0A125RA09 A0A2Z5RE76 B3P3R7 A0A0N8CDK3 A0A1A9ZZJ7 A0A1A9WJB9 A0A1Y1LUA6 R4FQA8 C9VXR9 A0A0P5CHV2 A0A1B6EXE0 T1PDA4 E0VKP4 A0A1B6CK57 A0A0A9W354 P83968 P83967 P83969 A0A1L8E4X1 A0A1B0D7E8 A0A151JMQ0 A0A1W4XH28 A0A1W4XWB9 A0A2Z4G633 A0A151I4W0 K7J803 X5CT98 A0A195FX88 A0A2R4GMB1 A0A158NXU2 A0A0P5CGV3 A0A3G1SUZ3 A0A151WIN4 A0A0P5CHD6 A0A0N8A945 A0A1L8E5H6 Q7Q7K6 A0A0P5EGX5 A0A2M4ATG0 A0A0H4IV48 Q5RLJ4 V5GR51 A0A023J916 D6WF19 A0A023VZM4 C8CD00 A0A0P5DTY5 A0A026X3P0 N6TH73 A0A1B1UZE0
PDB
5CE3
E-value=0,
Score=1999
Ontologies
PATHWAY
GO
PANTHER
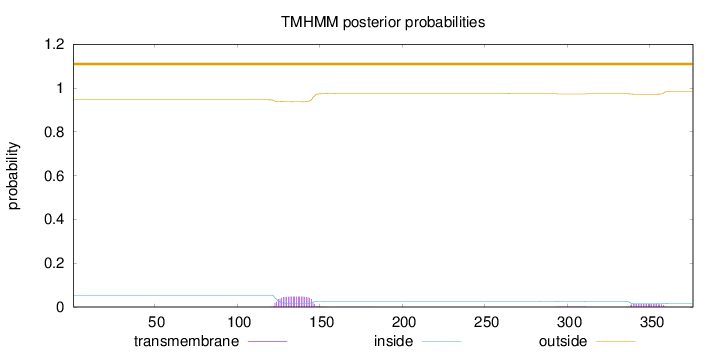
Topology
Subcellular location
Length:
376
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.35323
Exp number, first 60 AAs:
0.00115
Total prob of N-in:
0.05368
outside
1 - 376
Population Genetic Test Statistics
Pi
4.820552
Theta
6.298703
Tajima's D
-0.374478
CLR
0.495425
CSRT
0.28243587820609
Interpretation
Uncertain
