Gene
KWMTBOMO04834
Pre Gene Modal
BGIBMGA009927
Annotation
neuropeptide_receptor_B1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.566
Sequence
CDS
ATGAACGACCCGGAGCGCATCGCCCGCGACCTTCTCCTCTGCGAAGAGTTCAACAATCGGACCTTGCCACCGCCAGGATTGTACTGCGAGGGGACGTTCGACCGCTGGCTGTGTTGGCCTCACACGCCCGCCAACAGCACCGCCTACGGTTCCTGTCCTGAGTTCGTTCCCGGATTCAGACCTGATCTGTTGGCGCACAAGGAATGCACTGCGAACGGAACCTGGTACAAGCACCCGGAAACCGGCCTTCCCTGGTCCAACTACACCACCTGCGTCGTCGAGGAGGATGACGTGAACCACATCATCGTGGTGTACGAGGCGGGCTACAGCGTGTCGCTGGTCGCCTTGCTGCTCTCCCTCGCCATCCTCTCGTACTTCAGGAGCCTGCGGTGCGCGCGCATCACGGTACACATGAACCTGTTCGGTTCGTTTGCGGTGAACAACGCGCTGTGGTTGGCGTGGTACGGGCTCGTGGTGCGCGACCCCACCACTCTGCAGGAGCCGCCCGTCTGGTGCTGTGCGCTTAATGCGGTGCTGCAGTACGCCATGCTCACCAACTACATGTGGATGCTGTGCGAGGGCATGTACCTGCACACGGTGCTGGTCAGTGCCTTCATCTCGGAGCGACGGCTCGTGCGCGCACTGGTGGCGGCGGGTTGGATGCTGCCACTCCCCTGCATCCTGATCTACGCCACGCGACGAGCACTCGACGGTGACCCGCTTTGCTGGGCCGAGGAGCCTGAATCGCGACCCGAGCTCGCCGTGCCGGTGGGCTTGGCCGTTCTGCTTAACCTGTGCTTCTTGTGCAACATCGTGCGCGTGCTGTGTACCAAACTGAGGGCGGGAGGCGCGGCAGGGGGGGCGGCCCGGCCGTCGGCCACCGCTTTGCACGCCCTCCGTGCCACCTGCCTGCTGGCCCCGCTGCTCGGCCTGCAGTACCTGCTGATGCCGTTTCGGCCCGCCAACACGGTGAGCTGGTGGCGCGCCTATGAGTACGCAACAGCGGCGGCGACCTCGCTGCAGGGTCTGTGCGTGGCGGTGCTGTACTGCTTCTGCAACGGCGAGGTGCTGGCACAGCTGCGGCGCCGGTGGCGCGCGCTCACGTTCCGGCCGAGAGCCAATTCCACGACAGCCACGACGGTTTCGTTCGTGCGGTCGGCGGCAACGGCCGGCGGCGAGGACAAGGTGTGA
Protein
MNDPERIARDLLLCEEFNNRTLPPPGLYCEGTFDRWLCWPHTPANSTAYGSCPEFVPGFRPDLLAHKECTANGTWYKHPETGLPWSNYTTCVVEEDDVNHIIVVYEAGYSVSLVALLLSLAILSYFRSLRCARITVHMNLFGSFAVNNALWLAWYGLVVRDPTTLQEPPVWCCALNAVLQYAMLTNYMWMLCEGMYLHTVLVSAFISERRLVRALVAAGWMLPLPCILIYATRRALDGDPLCWAEEPESRPELAVPVGLAVLLNLCFLCNIVRVLCTKLRAGGAAGGAARPSATALHALRATCLLAPLLGLQYLLMPFRPANTVSWWRAYEYATAAATSLQGLCVAVLYCFCNGEVLAQLRRRWRALTFRPRANSTTATTVSFVRSAATAGGEDKV
Summary
Similarity
Belongs to the G-protein coupled receptor 2 family.
Uniprot
B3XXP9
H9JK76
A0A212F398
A0A194RD82
A0A0S1YDC1
A0A194PS72
+ More
A0A2W1BHD2 A0A2H1WBE8 A0A1L8DJZ5 A0A1B6FEL9 U3U4H7 E0VT86 B4HQH4 A0A0J9RBT4 A1Z9B7 A0A3B0JS22 A0A024E293 A0A0M4EIX2 A0A0B4KF12 K7J018 A0A1L8DJU9 A0A1B6DAB7 A0A3B0J063 A0A2U9PFW0 B3MGX1 A0A1B6HNS4 Q293B3 A0A1B6LXJ3 A0A0K8VHA2 A0A1B6GBP0 B4P4J0 A0A034WGP1 A0A0K8UB56 B4GBY6 A0A0B4KEN5 B3NRM6 B4N613 A0A1I8M1A3 A0A3B0JF33 B4J5C4 A0A3B0JKD0 B4LLZ0 A0A1Q3FRE0 B4KLM4 W8AQB8 A0A3Q0IXJ0 A0A0K8VAV3 V5UZ27 A0A0K8W8K9 A0A0K8WC78 A0A0Q9W2C8 K9KYK1 A0A1W4XSW4 A0A2S2NU23 A0A1Y1M8Y7 A0A088A7A3 W8B0D7 A0A2M4ANI3 V5UZV0 A0A1Q3FQS0 A0A2M4CY09 A0A1L8DJN0 A0A2S2QSG2 A0A2H8THG7 A0A1W4UAF3 A0A3L8DW89 A0A336M5R0 A0A2M4BPS8 B0X5H4 A0A0C9RV18 A0A182GV21 A0A182WGK6 W5JAX3 A0A182LTM8 A0A182QGL0 B4QE19 A0A1J1IVL4 A0A182Y0Q2 A0A182F2W2 A0A182NBS8 T1E209 A0A0L7QNH9 A0A1J1IXK7 A0A182J2M7 J9K361 T1I0J4 A0A067R9A4 A0A1W4XHU8 A0A151WMD3 A0A2A3EEV2 A0A195B197 E2ADW1 A0A026WP15 N6U0C2 A0A182KU03 A0A345BTT1 E9HF82 A0A182HRV8 A0A182TWX0
A0A2W1BHD2 A0A2H1WBE8 A0A1L8DJZ5 A0A1B6FEL9 U3U4H7 E0VT86 B4HQH4 A0A0J9RBT4 A1Z9B7 A0A3B0JS22 A0A024E293 A0A0M4EIX2 A0A0B4KF12 K7J018 A0A1L8DJU9 A0A1B6DAB7 A0A3B0J063 A0A2U9PFW0 B3MGX1 A0A1B6HNS4 Q293B3 A0A1B6LXJ3 A0A0K8VHA2 A0A1B6GBP0 B4P4J0 A0A034WGP1 A0A0K8UB56 B4GBY6 A0A0B4KEN5 B3NRM6 B4N613 A0A1I8M1A3 A0A3B0JF33 B4J5C4 A0A3B0JKD0 B4LLZ0 A0A1Q3FRE0 B4KLM4 W8AQB8 A0A3Q0IXJ0 A0A0K8VAV3 V5UZ27 A0A0K8W8K9 A0A0K8WC78 A0A0Q9W2C8 K9KYK1 A0A1W4XSW4 A0A2S2NU23 A0A1Y1M8Y7 A0A088A7A3 W8B0D7 A0A2M4ANI3 V5UZV0 A0A1Q3FQS0 A0A2M4CY09 A0A1L8DJN0 A0A2S2QSG2 A0A2H8THG7 A0A1W4UAF3 A0A3L8DW89 A0A336M5R0 A0A2M4BPS8 B0X5H4 A0A0C9RV18 A0A182GV21 A0A182WGK6 W5JAX3 A0A182LTM8 A0A182QGL0 B4QE19 A0A1J1IVL4 A0A182Y0Q2 A0A182F2W2 A0A182NBS8 T1E209 A0A0L7QNH9 A0A1J1IXK7 A0A182J2M7 J9K361 T1I0J4 A0A067R9A4 A0A1W4XHU8 A0A151WMD3 A0A2A3EEV2 A0A195B197 E2ADW1 A0A026WP15 N6U0C2 A0A182KU03 A0A345BTT1 E9HF82 A0A182HRV8 A0A182TWX0
Pubmed
18725956
19121390
22118469
26354079
28756777
23932938
+ More
20566863 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20075255 15632085 17550304 25348373 25315136 24495485 24312424 23209727 28004739 30249741 26483478 20920257 23761445 25244985 24330624 24845553 20798317 24508170 23537049 20966253 30022930 21292972
20566863 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20075255 15632085 17550304 25348373 25315136 24495485 24312424 23209727 28004739 30249741 26483478 20920257 23761445 25244985 24330624 24845553 20798317 24508170 23537049 20966253 30022930 21292972
EMBL
AB330457
BAG68435.1
BABH01026770
BABH01026771
AGBW02010600
OWR48204.1
+ More
KQ460367 KPJ15429.1 KT031043 RSAL01000018 ALM88341.1 RVE52858.1 KQ459595 KPI95813.1 KZ150126 PZC73115.1 ODYU01007534 SOQ50388.1 GFDF01007420 JAV06664.1 GECZ01021215 JAS48554.1 AB817334 BAO01101.1 DS235760 EEB16592.1 CH480816 EDW47709.1 CM002911 KMY93468.1 AE013599 AAN16138.1 OUUW01000001 SPP73908.1 BT150448 AHZ64222.1 CP012524 ALC41130.1 AGB93483.1 GFDF01007346 JAV06738.1 GEDC01014657 JAS22641.1 SPP73907.1 MG550228 AWT50662.1 CH902619 EDV37889.2 GECU01031372 JAS76334.1 CM000071 EAL24698.3 GEBQ01025341 GEBQ01011586 JAT14636.1 JAT28391.1 GDHF01014081 JAI38233.1 GECZ01010056 JAS59713.1 CM000158 EDW90629.2 GAKP01005652 JAC53300.1 GDHF01028729 GDHF01017767 JAI23585.1 JAI34547.1 CH479181 EDW31365.1 AGB93482.1 CH954179 EDV56178.2 CH964154 EDW79802.2 SPP73910.1 CH916367 EDW01766.1 SPP73909.1 CH940648 EDW59910.2 GFDL01005062 JAV29983.1 CH933808 EDW08660.2 GAMC01019887 JAB86668.1 GDHF01016338 JAI35976.1 KC660149 AHB86317.1 GDHF01004955 JAI47359.1 GDHF01003617 JAI48697.1 KRF79149.1 JQ045343 AEU12191.1 GGMR01008082 MBY20701.1 GEZM01037623 JAV82021.1 GAMC01019884 JAB86671.1 GGFK01008971 MBW42292.1 KC660150 AHB86318.1 GFDL01005074 JAV29971.1 GGFL01006036 MBW70214.1 GFDF01007421 JAV06663.1 GGMS01011400 MBY80603.1 GFXV01001694 MBW13499.1 QOIP01000003 RLU24626.1 UFQT01000565 SSX25360.1 GGFJ01005871 MBW55012.1 DS232379 EDS40859.1 GBYB01011516 JAG81283.1 JXUM01018724 JXUM01018725 JXUM01018726 JXUM01018727 JXUM01018728 KQ560520 KXJ82009.1 ADMH02002010 ETN59995.1 AXCM01002423 AXCN02000686 CM000362 EDX06913.1 CVRI01000063 CRL04307.1 GALA01001620 JAA93232.1 KQ414851 KOC60173.1 CRL04308.1 ABLF02035644 ABLF02035645 ABLF02035649 ABLF02035650 ACPB03022822 KK852804 KDR16174.1 KQ982944 KYQ48961.1 KZ288279 PBC29696.1 KQ976691 KYM77974.1 GL438820 EFN68473.1 KK107139 EZA57760.1 APGK01044279 APGK01044280 APGK01044281 KB741022 ENN74985.1 MH331892 AXF54360.1 GL732634 EFX69566.1 APCN01001764
KQ460367 KPJ15429.1 KT031043 RSAL01000018 ALM88341.1 RVE52858.1 KQ459595 KPI95813.1 KZ150126 PZC73115.1 ODYU01007534 SOQ50388.1 GFDF01007420 JAV06664.1 GECZ01021215 JAS48554.1 AB817334 BAO01101.1 DS235760 EEB16592.1 CH480816 EDW47709.1 CM002911 KMY93468.1 AE013599 AAN16138.1 OUUW01000001 SPP73908.1 BT150448 AHZ64222.1 CP012524 ALC41130.1 AGB93483.1 GFDF01007346 JAV06738.1 GEDC01014657 JAS22641.1 SPP73907.1 MG550228 AWT50662.1 CH902619 EDV37889.2 GECU01031372 JAS76334.1 CM000071 EAL24698.3 GEBQ01025341 GEBQ01011586 JAT14636.1 JAT28391.1 GDHF01014081 JAI38233.1 GECZ01010056 JAS59713.1 CM000158 EDW90629.2 GAKP01005652 JAC53300.1 GDHF01028729 GDHF01017767 JAI23585.1 JAI34547.1 CH479181 EDW31365.1 AGB93482.1 CH954179 EDV56178.2 CH964154 EDW79802.2 SPP73910.1 CH916367 EDW01766.1 SPP73909.1 CH940648 EDW59910.2 GFDL01005062 JAV29983.1 CH933808 EDW08660.2 GAMC01019887 JAB86668.1 GDHF01016338 JAI35976.1 KC660149 AHB86317.1 GDHF01004955 JAI47359.1 GDHF01003617 JAI48697.1 KRF79149.1 JQ045343 AEU12191.1 GGMR01008082 MBY20701.1 GEZM01037623 JAV82021.1 GAMC01019884 JAB86671.1 GGFK01008971 MBW42292.1 KC660150 AHB86318.1 GFDL01005074 JAV29971.1 GGFL01006036 MBW70214.1 GFDF01007421 JAV06663.1 GGMS01011400 MBY80603.1 GFXV01001694 MBW13499.1 QOIP01000003 RLU24626.1 UFQT01000565 SSX25360.1 GGFJ01005871 MBW55012.1 DS232379 EDS40859.1 GBYB01011516 JAG81283.1 JXUM01018724 JXUM01018725 JXUM01018726 JXUM01018727 JXUM01018728 KQ560520 KXJ82009.1 ADMH02002010 ETN59995.1 AXCM01002423 AXCN02000686 CM000362 EDX06913.1 CVRI01000063 CRL04307.1 GALA01001620 JAA93232.1 KQ414851 KOC60173.1 CRL04308.1 ABLF02035644 ABLF02035645 ABLF02035649 ABLF02035650 ACPB03022822 KK852804 KDR16174.1 KQ982944 KYQ48961.1 KZ288279 PBC29696.1 KQ976691 KYM77974.1 GL438820 EFN68473.1 KK107139 EZA57760.1 APGK01044279 APGK01044280 APGK01044281 KB741022 ENN74985.1 MH331892 AXF54360.1 GL732634 EFX69566.1 APCN01001764
Proteomes
UP000005204
UP000007151
UP000053240
UP000283053
UP000053268
UP000009046
+ More
UP000001292 UP000000803 UP000268350 UP000092553 UP000002358 UP000007801 UP000001819 UP000002282 UP000008744 UP000008711 UP000007798 UP000095301 UP000001070 UP000008792 UP000009192 UP000079169 UP000192223 UP000005203 UP000192221 UP000279307 UP000002320 UP000069940 UP000249989 UP000075920 UP000000673 UP000075883 UP000075886 UP000000304 UP000183832 UP000076408 UP000069272 UP000075884 UP000053825 UP000075880 UP000007819 UP000015103 UP000027135 UP000075809 UP000242457 UP000078540 UP000000311 UP000053097 UP000019118 UP000075882 UP000000305 UP000075840 UP000075902
UP000001292 UP000000803 UP000268350 UP000092553 UP000002358 UP000007801 UP000001819 UP000002282 UP000008744 UP000008711 UP000007798 UP000095301 UP000001070 UP000008792 UP000009192 UP000079169 UP000192223 UP000005203 UP000192221 UP000279307 UP000002320 UP000069940 UP000249989 UP000075920 UP000000673 UP000075883 UP000075886 UP000000304 UP000183832 UP000076408 UP000069272 UP000075884 UP000053825 UP000075880 UP000007819 UP000015103 UP000027135 UP000075809 UP000242457 UP000078540 UP000000311 UP000053097 UP000019118 UP000075882 UP000000305 UP000075840 UP000075902
Interpro
SUPFAM
SSF111418
SSF111418
Gene 3D
ProteinModelPortal
B3XXP9
H9JK76
A0A212F398
A0A194RD82
A0A0S1YDC1
A0A194PS72
+ More
A0A2W1BHD2 A0A2H1WBE8 A0A1L8DJZ5 A0A1B6FEL9 U3U4H7 E0VT86 B4HQH4 A0A0J9RBT4 A1Z9B7 A0A3B0JS22 A0A024E293 A0A0M4EIX2 A0A0B4KF12 K7J018 A0A1L8DJU9 A0A1B6DAB7 A0A3B0J063 A0A2U9PFW0 B3MGX1 A0A1B6HNS4 Q293B3 A0A1B6LXJ3 A0A0K8VHA2 A0A1B6GBP0 B4P4J0 A0A034WGP1 A0A0K8UB56 B4GBY6 A0A0B4KEN5 B3NRM6 B4N613 A0A1I8M1A3 A0A3B0JF33 B4J5C4 A0A3B0JKD0 B4LLZ0 A0A1Q3FRE0 B4KLM4 W8AQB8 A0A3Q0IXJ0 A0A0K8VAV3 V5UZ27 A0A0K8W8K9 A0A0K8WC78 A0A0Q9W2C8 K9KYK1 A0A1W4XSW4 A0A2S2NU23 A0A1Y1M8Y7 A0A088A7A3 W8B0D7 A0A2M4ANI3 V5UZV0 A0A1Q3FQS0 A0A2M4CY09 A0A1L8DJN0 A0A2S2QSG2 A0A2H8THG7 A0A1W4UAF3 A0A3L8DW89 A0A336M5R0 A0A2M4BPS8 B0X5H4 A0A0C9RV18 A0A182GV21 A0A182WGK6 W5JAX3 A0A182LTM8 A0A182QGL0 B4QE19 A0A1J1IVL4 A0A182Y0Q2 A0A182F2W2 A0A182NBS8 T1E209 A0A0L7QNH9 A0A1J1IXK7 A0A182J2M7 J9K361 T1I0J4 A0A067R9A4 A0A1W4XHU8 A0A151WMD3 A0A2A3EEV2 A0A195B197 E2ADW1 A0A026WP15 N6U0C2 A0A182KU03 A0A345BTT1 E9HF82 A0A182HRV8 A0A182TWX0
A0A2W1BHD2 A0A2H1WBE8 A0A1L8DJZ5 A0A1B6FEL9 U3U4H7 E0VT86 B4HQH4 A0A0J9RBT4 A1Z9B7 A0A3B0JS22 A0A024E293 A0A0M4EIX2 A0A0B4KF12 K7J018 A0A1L8DJU9 A0A1B6DAB7 A0A3B0J063 A0A2U9PFW0 B3MGX1 A0A1B6HNS4 Q293B3 A0A1B6LXJ3 A0A0K8VHA2 A0A1B6GBP0 B4P4J0 A0A034WGP1 A0A0K8UB56 B4GBY6 A0A0B4KEN5 B3NRM6 B4N613 A0A1I8M1A3 A0A3B0JF33 B4J5C4 A0A3B0JKD0 B4LLZ0 A0A1Q3FRE0 B4KLM4 W8AQB8 A0A3Q0IXJ0 A0A0K8VAV3 V5UZ27 A0A0K8W8K9 A0A0K8WC78 A0A0Q9W2C8 K9KYK1 A0A1W4XSW4 A0A2S2NU23 A0A1Y1M8Y7 A0A088A7A3 W8B0D7 A0A2M4ANI3 V5UZV0 A0A1Q3FQS0 A0A2M4CY09 A0A1L8DJN0 A0A2S2QSG2 A0A2H8THG7 A0A1W4UAF3 A0A3L8DW89 A0A336M5R0 A0A2M4BPS8 B0X5H4 A0A0C9RV18 A0A182GV21 A0A182WGK6 W5JAX3 A0A182LTM8 A0A182QGL0 B4QE19 A0A1J1IVL4 A0A182Y0Q2 A0A182F2W2 A0A182NBS8 T1E209 A0A0L7QNH9 A0A1J1IXK7 A0A182J2M7 J9K361 T1I0J4 A0A067R9A4 A0A1W4XHU8 A0A151WMD3 A0A2A3EEV2 A0A195B197 E2ADW1 A0A026WP15 N6U0C2 A0A182KU03 A0A345BTT1 E9HF82 A0A182HRV8 A0A182TWX0
PDB
5UZ7
E-value=5.85238e-54,
Score=533
Ontologies
GO
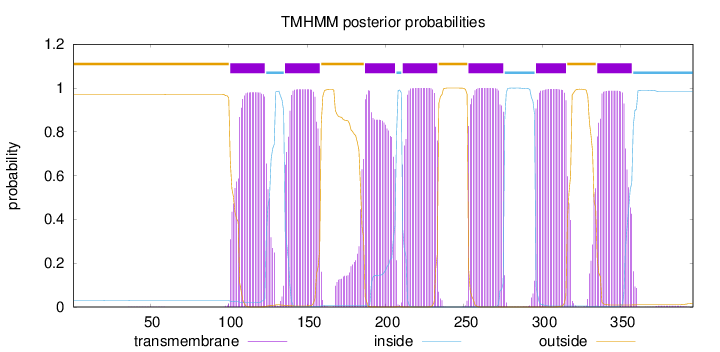
Topology
Length:
396
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
153.02185
Exp number, first 60 AAs:
0.00694
Total prob of N-in:
0.03028
outside
1 - 100
TMhelix
101 - 123
inside
124 - 135
TMhelix
136 - 158
outside
159 - 186
TMhelix
187 - 206
inside
207 - 210
TMhelix
211 - 233
outside
234 - 252
TMhelix
253 - 275
inside
276 - 295
TMhelix
296 - 315
outside
316 - 334
TMhelix
335 - 357
inside
358 - 396
Population Genetic Test Statistics
Pi
14.242946
Theta
13.122262
Tajima's D
-0.779586
CLR
0.623886
CSRT
0.182090895455227
Interpretation
Uncertain
