Gene
KWMTBOMO04832
Pre Gene Modal
BGIBMGA009925
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.037 Extracellular Reliability : 1.346
Sequence
CDS
ATGAGGTGCCTGGTCGCCTGCTCCCCCATGGCCGATGGAGCCGCGTATGATTACGTGGTGGTAGGAGCTGGGGCTGCGGGCGCCGCGGCCGCCTCTAGGTTAGCATTGAAAGGTTTCGATGTTATGCTTATAGAAGCCGGAGGCGATCCAAGCTTCCTATCTACGATACCGATGGCGTCGTTGGGTCTCCTTGGCTCCTCACTGGATTGGCAGTACAAGACTATTCCAAACAACATATCCTGTCTCTCTTCAATTGGTGAACAGTGTAGATTCAGTCGTGGAAAGAGCTTAGGAGGAACCACCGCTATCAATCACATGCTGTACATAAGAGGAAATAGATATGATTACGACAGAATGAACATCTCAGGGTGGACATGGAAGGACTTAGAGCCGTATTTCCTTCGATACGAAGGTCTTCAAATTTTAGACCAGTATCCAGCCAGTTCAAGGAAATATCATAATAGTAATGGTACTATGAAATTGGAGTATTTTGATGATCCGCGTAACCCATGGCACATGAGAACTGTTGAGGGGTTAAAGCAGCTAAATATACCTTATAATAAGGATTTGAACGGAGAGTATCAAATAGGAGTTACAAAAGTAGCTGGTTATGTATATAAAGGAGAGAGGATGAGTACCGCGCGGGGCTACCTAGCAAGGAATGATGTTAAGAAAAATCTTAAAGTTGCAAAGAATGCTTTTTGTACTGAAGTAGTTATTGATGACGACAATATTGCTCGAGGAGTGACAGTTGTCCAAAATCTACAAAAAGTTACAATATATGCAAGAATCGAAATTATACTTAGTGCTGGAGCGATTGGAACCCCTCAGATATTGATGCAGTCAGGTGTCGGTCCAGCTGACCACTTACAAGAAATGGGCATATCAGTACGATCAAACCTTCCCGTCGGTAATGATATGAGTGACCATCAGCTACCAGTAGTCATCGTCAAAGTAGACCACGGAGGAATAGTAGACAGCCTCGTTGGACTGACTTCTAAAGCACCCCAAGTATTACAGTACTTGGCCTCCAGGAGCGGACCACTAGCATCTAATAGCCTACAAGATATAGTGACATTGATAAACACTAACTGTTACGATTTTGATCTTCGTCAGTATTCAGGTAATAGTAGTCGGTGCGAGCTCGCTGATATGCAGATTATTCAGTCGTACATAGACAAAGGATTGGTGGCTTTAGCAAAGCCTTTGGTTAAGCAGGCTATTGGTTTCAATGACCAAGTCTTAGACCAAATTGAGAAGGCGAACGAGAACCATGGCTTGATTGTTTTCTCGCCTATGGTTCTGCAGCCATATTCTCGTGGCACCGTCCGCCTCGCCAGTATTGATCCACTGCAACCACCAGCAATCTTCGCTAATTATCTTGGAGATGAACGAGACGTCGAACAAATGGTAAAATCTATCACTTTCTTGGAGCACCTGATGAAAACTCAGATCTTCAAAAAACACAAAGCCTCAATCTTACATTTGAATCTACCTGGATGTCCAGCCTATAAATGTGGAAGGGTAGAGTATTGGAGATGCTACGCCAGACATATGACTTATTCTGGATACCATGCTGTAGGGACCTGTGCGCTGACCAGGGTGGTAGATGAACAACTCCGTGTGTATGGTGTAAAGAACTTAAGAGTGGCCGATTTGAGTGTACTGAAAAATATTATAAGAGGAAACACAGCATCTGTTTCTATAGCTATTGGAGAGAGAATTGTGGATTTTTTAACTGAGGAAGATAATCCATAG
Protein
MRCLVACSPMADGAAYDYVVVGAGAAGAAAASRLALKGFDVMLIEAGGDPSFLSTIPMASLGLLGSSLDWQYKTIPNNISCLSSIGEQCRFSRGKSLGGTTAINHMLYIRGNRYDYDRMNISGWTWKDLEPYFLRYEGLQILDQYPASSRKYHNSNGTMKLEYFDDPRNPWHMRTVEGLKQLNIPYNKDLNGEYQIGVTKVAGYVYKGERMSTARGYLARNDVKKNLKVAKNAFCTEVVIDDDNIARGVTVVQNLQKVTIYARIEIILSAGAIGTPQILMQSGVGPADHLQEMGISVRSNLPVGNDMSDHQLPVVIVKVDHGGIVDSLVGLTSKAPQVLQYLASRSGPLASNSLQDIVTLINTNCYDFDLRQYSGNSSRCELADMQIIQSYIDKGLVALAKPLVKQAIGFNDQVLDQIEKANENHGLIVFSPMVLQPYSRGTVRLASIDPLQPPAIFANYLGDERDVEQMVKSITFLEHLMKTQIFKKHKASILHLNLPGCPAYKCGRVEYWRCYARHMTYSGYHAVGTCALTRVVDEQLRVYGVKNLRVADLSVLKNIIRGNTASVSIAIGERIVDFLTEEDNP
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9JK74
H9JK73
A0A0L7LH02
A0A2W1BN46
A0A212FEK7
A0A212F392
+ More
A0A2A4IZN2 A0A0L7LNY9 A0A3S2LSE6 A0A0L7KY32 A0A3S2P1X7 A0A2K8FTL4 A0A1B6LHN9 A0A1S6J0X2 A0A182FZT3 E9G975 A0A026WCZ8 A0A0P5E9N8 A0A3L8D7W0 E2ASE9 A0A1L8E0M7 A0A0C9R3W9 K7J281 A0A1L8E0S3 A0A0T6BDD9 A0A2S2QPB6 A0A2P8YRH6 A0A0P4Z1D7 A0A2H8TYC1 T1J5G0 D2A3N1 A0A2S2P6F1 A0A2J7RBI7 A0A151K0E9 A0A162CBJ1 A0A088AJV0 A0A154PBR8 A0A0P5TTW1 A0A0M9AA24 E9IKG5 A0A210Q328 A0A0J7KLX4 E9G974 A0A2P8ZKS2 F4WR74 A0A0P5BAV0 A0A0L7RJZ7 J9JM93 A0A3L8D7B4 A0A151J323 A0A1W4VFW5 A0A0C9RDB5 T1ILS9 A0A1B6JVY6 A0A1B6K5Y4 A0A0J7K353 A0A2J7PXM4 A0A210Q8G7 A0A0P5C063 A0A084VW82 A0A158NUR3 K7J280 A0A151WNR9 A0A226D4L8 A0A2K8FTP1 A0A0N8A6J9 E0VUA7 A0A0P5CC33 B3NUX3 A0A0A1WEA8 A0A2H1WZL6 A0A1L8E0E7 A0A034VNI0 A0A2A4IYS9 E2BJJ0 A0A154PBS2 A0A2P8ZKR5 A0A0P5J4G4 A0A0P5NHU5 A0A1L8DZX6 A0A1B6MBY6 A0A0P5L7L0 A0A0P5CQE2 A0A0A1WXX2 A0A0P6FZX5 A0A0P4YIW2 A0A0P6F7Y1 A0A1B6EYD4 A0A0P5S3M2 A0A232EPA2 A0A0P5RGK1 A0A151K093 A0A310SXN7 A0A0P5JE16 A0A0P5JVR4 A0A1D2MH79 A0A0P6FAU5 A0A0N8AWF9 A0A151IVV0 A0A310SSQ6
A0A2A4IZN2 A0A0L7LNY9 A0A3S2LSE6 A0A0L7KY32 A0A3S2P1X7 A0A2K8FTL4 A0A1B6LHN9 A0A1S6J0X2 A0A182FZT3 E9G975 A0A026WCZ8 A0A0P5E9N8 A0A3L8D7W0 E2ASE9 A0A1L8E0M7 A0A0C9R3W9 K7J281 A0A1L8E0S3 A0A0T6BDD9 A0A2S2QPB6 A0A2P8YRH6 A0A0P4Z1D7 A0A2H8TYC1 T1J5G0 D2A3N1 A0A2S2P6F1 A0A2J7RBI7 A0A151K0E9 A0A162CBJ1 A0A088AJV0 A0A154PBR8 A0A0P5TTW1 A0A0M9AA24 E9IKG5 A0A210Q328 A0A0J7KLX4 E9G974 A0A2P8ZKS2 F4WR74 A0A0P5BAV0 A0A0L7RJZ7 J9JM93 A0A3L8D7B4 A0A151J323 A0A1W4VFW5 A0A0C9RDB5 T1ILS9 A0A1B6JVY6 A0A1B6K5Y4 A0A0J7K353 A0A2J7PXM4 A0A210Q8G7 A0A0P5C063 A0A084VW82 A0A158NUR3 K7J280 A0A151WNR9 A0A226D4L8 A0A2K8FTP1 A0A0N8A6J9 E0VUA7 A0A0P5CC33 B3NUX3 A0A0A1WEA8 A0A2H1WZL6 A0A1L8E0E7 A0A034VNI0 A0A2A4IYS9 E2BJJ0 A0A154PBS2 A0A2P8ZKR5 A0A0P5J4G4 A0A0P5NHU5 A0A1L8DZX6 A0A1B6MBY6 A0A0P5L7L0 A0A0P5CQE2 A0A0A1WXX2 A0A0P6FZX5 A0A0P4YIW2 A0A0P6F7Y1 A0A1B6EYD4 A0A0P5S3M2 A0A232EPA2 A0A0P5RGK1 A0A151K093 A0A310SXN7 A0A0P5JE16 A0A0P5JVR4 A0A1D2MH79 A0A0P6FAU5 A0A0N8AWF9 A0A151IVV0 A0A310SSQ6
Pubmed
EMBL
BABH01026760
BABH01026761
JTDY01001166
KOB74675.1
KZ150126
PZC73113.1
+ More
AGBW02008930 OWR52172.1 AGBW02010600 OWR48201.1 NWSH01004950 PCG64563.1 JTDY01000443 KOB77147.1 RSAL01000892 RSAL01000018 RVE41068.1 RVE52863.1 JTDY01004610 KOB67954.1 RSAL01000051 RVE50264.1 KY618825 AQW43011.1 GEBQ01016745 JAT23232.1 KU764422 AQS60669.1 GL732535 EFX84138.1 KK107261 EZA53937.1 GDIP01146283 JAJ77119.1 QOIP01000012 RLU16172.1 GL442298 EFN63615.1 GFDF01001806 JAV12278.1 GBYB01002620 JAG72387.1 AAZX01008029 GFDF01001756 JAV12328.1 LJIG01001624 KRT85316.1 GGMS01010167 MBY79370.1 PYGN01000411 PSN46845.1 GDIP01219778 JAJ03624.1 GFXV01007441 MBW19246.1 JH431861 KQ971348 EFA05531.1 GGMR01012313 MBY24932.1 NEVH01005904 PNF38181.1 KQ981296 KYN43080.1 LRGB01001036 KZS13782.1 KQ434869 KZC09326.1 GDIP01121629 JAL82085.1 KQ435704 KOX80305.1 GL763984 EFZ18935.1 NEDP02005158 OWF43153.1 LBMM01005695 KMQ91287.1 EFX84139.1 PYGN01000027 PSN57097.1 GL888284 EGI63349.1 GDIP01187565 JAJ35837.1 KQ414579 KOC71128.1 ABLF02037928 RLU16179.1 KQ980314 KYN16692.1 GBYB01011122 JAG80889.1 JH430884 GECU01004364 JAT03343.1 GECU01001143 JAT06564.1 LBMM01015561 KMQ84762.1 NEVH01020853 PNF21087.1 NEDP02004628 OWF45028.1 GDIP01177544 JAJ45858.1 ATLV01017501 KE525172 KFB42226.1 ADTU01026685 AAZX01001588 KQ982905 KYQ49514.1 LNIX01000033 OXA40505.1 KY618826 AQW43012.1 GDIP01177545 JAJ45857.1 DS235784 EEB16963.1 GDIP01177546 JAJ45856.1 CH954180 EDV47071.1 GBXI01017045 GBXI01016813 GBXI01008932 JAC97246.1 JAC97478.1 JAD05360.1 ODYU01012241 SOQ58442.1 GFDF01002042 JAV12042.1 GAKP01015844 JAC43108.1 NWSH01004668 PCG64789.1 GL448571 EFN84147.1 KZC09325.1 PSN57098.1 GDIQ01207189 JAK44536.1 GDIQ01162162 GDIQ01145021 GDIQ01143630 JAL06705.1 GFDF01002041 JAV12043.1 GEBQ01006583 JAT33394.1 GDIQ01200150 GDIQ01198855 JAK52870.1 GDIP01167184 JAJ56218.1 GBXI01010590 GBXI01000279 JAD03702.1 JAD14013.1 GDIQ01051491 JAN43246.1 GDIP01228120 JAI95281.1 GDIQ01051490 JAN43247.1 GECZ01026787 JAS42982.1 GDIQ01101823 JAL49903.1 NNAY01002979 OXU20190.1 GDIQ01101824 JAL49902.1 KYN43077.1 KQ759784 OAD62856.1 GDIQ01201409 JAK50316.1 GDIQ01198854 JAK52871.1 LJIJ01001277 ODM92293.1 GDIQ01051489 JAN43248.1 GDIP01238481 GDIQ01236187 GDIQ01234983 JAI84920.1 JAK15538.1 KQ980881 KYN11981.1 OAD62857.1
AGBW02008930 OWR52172.1 AGBW02010600 OWR48201.1 NWSH01004950 PCG64563.1 JTDY01000443 KOB77147.1 RSAL01000892 RSAL01000018 RVE41068.1 RVE52863.1 JTDY01004610 KOB67954.1 RSAL01000051 RVE50264.1 KY618825 AQW43011.1 GEBQ01016745 JAT23232.1 KU764422 AQS60669.1 GL732535 EFX84138.1 KK107261 EZA53937.1 GDIP01146283 JAJ77119.1 QOIP01000012 RLU16172.1 GL442298 EFN63615.1 GFDF01001806 JAV12278.1 GBYB01002620 JAG72387.1 AAZX01008029 GFDF01001756 JAV12328.1 LJIG01001624 KRT85316.1 GGMS01010167 MBY79370.1 PYGN01000411 PSN46845.1 GDIP01219778 JAJ03624.1 GFXV01007441 MBW19246.1 JH431861 KQ971348 EFA05531.1 GGMR01012313 MBY24932.1 NEVH01005904 PNF38181.1 KQ981296 KYN43080.1 LRGB01001036 KZS13782.1 KQ434869 KZC09326.1 GDIP01121629 JAL82085.1 KQ435704 KOX80305.1 GL763984 EFZ18935.1 NEDP02005158 OWF43153.1 LBMM01005695 KMQ91287.1 EFX84139.1 PYGN01000027 PSN57097.1 GL888284 EGI63349.1 GDIP01187565 JAJ35837.1 KQ414579 KOC71128.1 ABLF02037928 RLU16179.1 KQ980314 KYN16692.1 GBYB01011122 JAG80889.1 JH430884 GECU01004364 JAT03343.1 GECU01001143 JAT06564.1 LBMM01015561 KMQ84762.1 NEVH01020853 PNF21087.1 NEDP02004628 OWF45028.1 GDIP01177544 JAJ45858.1 ATLV01017501 KE525172 KFB42226.1 ADTU01026685 AAZX01001588 KQ982905 KYQ49514.1 LNIX01000033 OXA40505.1 KY618826 AQW43012.1 GDIP01177545 JAJ45857.1 DS235784 EEB16963.1 GDIP01177546 JAJ45856.1 CH954180 EDV47071.1 GBXI01017045 GBXI01016813 GBXI01008932 JAC97246.1 JAC97478.1 JAD05360.1 ODYU01012241 SOQ58442.1 GFDF01002042 JAV12042.1 GAKP01015844 JAC43108.1 NWSH01004668 PCG64789.1 GL448571 EFN84147.1 KZC09325.1 PSN57098.1 GDIQ01207189 JAK44536.1 GDIQ01162162 GDIQ01145021 GDIQ01143630 JAL06705.1 GFDF01002041 JAV12043.1 GEBQ01006583 JAT33394.1 GDIQ01200150 GDIQ01198855 JAK52870.1 GDIP01167184 JAJ56218.1 GBXI01010590 GBXI01000279 JAD03702.1 JAD14013.1 GDIQ01051491 JAN43246.1 GDIP01228120 JAI95281.1 GDIQ01051490 JAN43247.1 GECZ01026787 JAS42982.1 GDIQ01101823 JAL49903.1 NNAY01002979 OXU20190.1 GDIQ01101824 JAL49902.1 KYN43077.1 KQ759784 OAD62856.1 GDIQ01201409 JAK50316.1 GDIQ01198854 JAK52871.1 LJIJ01001277 ODM92293.1 GDIQ01051489 JAN43248.1 GDIP01238481 GDIQ01236187 GDIQ01234983 JAI84920.1 JAK15538.1 KQ980881 KYN11981.1 OAD62857.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000218220
UP000283053
UP000069272
+ More
UP000000305 UP000053097 UP000279307 UP000000311 UP000002358 UP000245037 UP000007266 UP000235965 UP000078541 UP000076858 UP000005203 UP000076502 UP000053105 UP000242188 UP000036403 UP000007755 UP000053825 UP000007819 UP000078492 UP000192221 UP000030765 UP000005205 UP000075809 UP000198287 UP000009046 UP000008711 UP000008237 UP000215335 UP000094527
UP000000305 UP000053097 UP000279307 UP000000311 UP000002358 UP000245037 UP000007266 UP000235965 UP000078541 UP000076858 UP000005203 UP000076502 UP000053105 UP000242188 UP000036403 UP000007755 UP000053825 UP000007819 UP000078492 UP000192221 UP000030765 UP000005205 UP000075809 UP000198287 UP000009046 UP000008711 UP000008237 UP000215335 UP000094527
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JK74
H9JK73
A0A0L7LH02
A0A2W1BN46
A0A212FEK7
A0A212F392
+ More
A0A2A4IZN2 A0A0L7LNY9 A0A3S2LSE6 A0A0L7KY32 A0A3S2P1X7 A0A2K8FTL4 A0A1B6LHN9 A0A1S6J0X2 A0A182FZT3 E9G975 A0A026WCZ8 A0A0P5E9N8 A0A3L8D7W0 E2ASE9 A0A1L8E0M7 A0A0C9R3W9 K7J281 A0A1L8E0S3 A0A0T6BDD9 A0A2S2QPB6 A0A2P8YRH6 A0A0P4Z1D7 A0A2H8TYC1 T1J5G0 D2A3N1 A0A2S2P6F1 A0A2J7RBI7 A0A151K0E9 A0A162CBJ1 A0A088AJV0 A0A154PBR8 A0A0P5TTW1 A0A0M9AA24 E9IKG5 A0A210Q328 A0A0J7KLX4 E9G974 A0A2P8ZKS2 F4WR74 A0A0P5BAV0 A0A0L7RJZ7 J9JM93 A0A3L8D7B4 A0A151J323 A0A1W4VFW5 A0A0C9RDB5 T1ILS9 A0A1B6JVY6 A0A1B6K5Y4 A0A0J7K353 A0A2J7PXM4 A0A210Q8G7 A0A0P5C063 A0A084VW82 A0A158NUR3 K7J280 A0A151WNR9 A0A226D4L8 A0A2K8FTP1 A0A0N8A6J9 E0VUA7 A0A0P5CC33 B3NUX3 A0A0A1WEA8 A0A2H1WZL6 A0A1L8E0E7 A0A034VNI0 A0A2A4IYS9 E2BJJ0 A0A154PBS2 A0A2P8ZKR5 A0A0P5J4G4 A0A0P5NHU5 A0A1L8DZX6 A0A1B6MBY6 A0A0P5L7L0 A0A0P5CQE2 A0A0A1WXX2 A0A0P6FZX5 A0A0P4YIW2 A0A0P6F7Y1 A0A1B6EYD4 A0A0P5S3M2 A0A232EPA2 A0A0P5RGK1 A0A151K093 A0A310SXN7 A0A0P5JE16 A0A0P5JVR4 A0A1D2MH79 A0A0P6FAU5 A0A0N8AWF9 A0A151IVV0 A0A310SSQ6
A0A2A4IZN2 A0A0L7LNY9 A0A3S2LSE6 A0A0L7KY32 A0A3S2P1X7 A0A2K8FTL4 A0A1B6LHN9 A0A1S6J0X2 A0A182FZT3 E9G975 A0A026WCZ8 A0A0P5E9N8 A0A3L8D7W0 E2ASE9 A0A1L8E0M7 A0A0C9R3W9 K7J281 A0A1L8E0S3 A0A0T6BDD9 A0A2S2QPB6 A0A2P8YRH6 A0A0P4Z1D7 A0A2H8TYC1 T1J5G0 D2A3N1 A0A2S2P6F1 A0A2J7RBI7 A0A151K0E9 A0A162CBJ1 A0A088AJV0 A0A154PBR8 A0A0P5TTW1 A0A0M9AA24 E9IKG5 A0A210Q328 A0A0J7KLX4 E9G974 A0A2P8ZKS2 F4WR74 A0A0P5BAV0 A0A0L7RJZ7 J9JM93 A0A3L8D7B4 A0A151J323 A0A1W4VFW5 A0A0C9RDB5 T1ILS9 A0A1B6JVY6 A0A1B6K5Y4 A0A0J7K353 A0A2J7PXM4 A0A210Q8G7 A0A0P5C063 A0A084VW82 A0A158NUR3 K7J280 A0A151WNR9 A0A226D4L8 A0A2K8FTP1 A0A0N8A6J9 E0VUA7 A0A0P5CC33 B3NUX3 A0A0A1WEA8 A0A2H1WZL6 A0A1L8E0E7 A0A034VNI0 A0A2A4IYS9 E2BJJ0 A0A154PBS2 A0A2P8ZKR5 A0A0P5J4G4 A0A0P5NHU5 A0A1L8DZX6 A0A1B6MBY6 A0A0P5L7L0 A0A0P5CQE2 A0A0A1WXX2 A0A0P6FZX5 A0A0P4YIW2 A0A0P6F7Y1 A0A1B6EYD4 A0A0P5S3M2 A0A232EPA2 A0A0P5RGK1 A0A151K093 A0A310SXN7 A0A0P5JE16 A0A0P5JVR4 A0A1D2MH79 A0A0P6FAU5 A0A0N8AWF9 A0A151IVV0 A0A310SSQ6
PDB
4YNU
E-value=1.11236e-35,
Score=377
Ontologies
GO
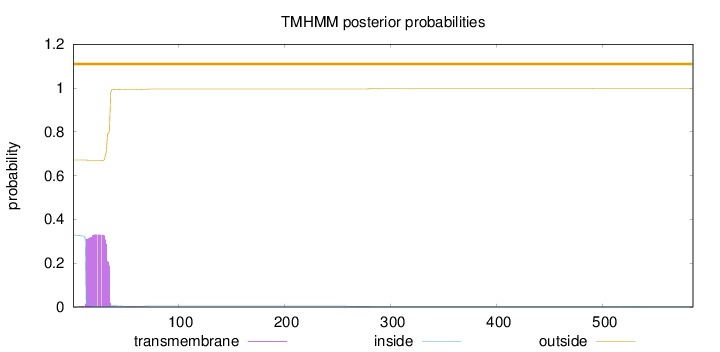
Topology
Length:
585
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.25918999999998
Exp number, first 60 AAs:
7.16612
Total prob of N-in:
0.32831
outside
1 - 585
Population Genetic Test Statistics
Pi
5.171972
Theta
7.508879
Tajima's D
-0.866909
CLR
1.016104
CSRT
0.16369181540923
Interpretation
Uncertain
