Gene
KWMTBOMO04827
Pre Gene Modal
BGIBMGA009825
Annotation
PREDICTED:_uncharacterized_protein_LOC101745460_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.403
Sequence
CDS
ATGTGGAGCCTGGTCGACAGTGGAGGAGCGGACGGCAAGTCTGCGCCGTGCTCCCGCGGCAAGCACTCCGCGACCTTACTAGGGGGCTTCGTGTACGTGCTGGGGGGGCGAGGGTCTGGAGGTTCCGTGCCGCTCAGAGATTTCTGGAGATATAGTCTTACAACAGGAAACTGGGAGAAGTTGGACTGTCGTGGAGAGCATCCTCCGTCTCTCCAAGAGCACACGGCGACCGCGCACGGGCACAGGATCTACGTGTTCGGCGGCGAGGCGGGGGCGCTCACCAACGAGACCCCGCTGTGGATATACGATACAGAGACACAAATCTGGCGCAAGCTTCCCGGCCAAGTGAACTACACTTCCTCCGTGCGTCGTCGCGCAAGGGACCCTCGTGGCGCTGTCAGTGGACCTCGAGGTCGGAGGGGACACACAGCTCATGCCCTTAAGGACTGTCTCCTCATTTATGGAGGGTACCAGGATTTGAAGGGGTCCACTAATGAACTCTGGGCGTTCCACTATGAATCCGAGTCGTGGCACCAAGTCCGGACGTGGAACTCCGGGCCGGCGCGCCACCGCCACGCCGCCGCGCTGCACGACGAGCGCCTGTACGTGCACGGCGGACAGTGCGACCTGCGGGACTGCGCCGACCTGTGGTTCTACGACACCATGTCGCGCGTGTGGCGGGAGGTGCGCGCGCGTGCGGGCCCGGCGCCGCGCAGTGGGCACGCGGCGGCGCGCGCTCGCGCTCACCTCTACATCTTCGGGGGAGACTCACATTCACACCCGCACAACGACCTGTGGCGGTTTCATTTTGCAACAGAAACATGGGAGCACATTCCTAATACAGTAAAGTGGCCCTCCCCTCGCACCGACACGTGCGCCATCCTCCTCCCGCGACGACCGCCTCGCGCTCCTCCCCGGTCCCCCCCGCTCGCCCACCCGACCCTGGCGCGGCCTCCTCCCCCACCTGGGAACATCCTTCGTGAAATCTCCAAGCTGTCTGCCTTTCACATCCGTCGCGCCGCTCGCTGCTCCTACACTGCCCTCGCCGGGAACAAGGACTCTACTGAAAGCTTAGTTCCATCCGAATCGACGCTTCCGAAGTCACAATCTGCCTTCCACATGGAGCGGCGCGGACAGTGCAGCCCGCGGGAAGGGGATGTACCCGACCTCACGCGTGACCCACTCTCCGTGCCCGACTTCGCGGACATGGTCACAACCCCCACTCGAACCACCAAGCTCGTCTACCTGGACTCTGAAGATGAAGCAAACGGGAAGAGAGACGTCACTGTTAAAATGGACAGCGAGACCACCGTGGTCATGCCCAAGTCAGCTTCTGTCCGGTTCACCAAAACTGTGACCATTACAGACGAAGAGGACACCGAGATGTCCACTTCAGACTACGCCAGCGCCGAGAAAGTGAACAGAATATCGGGAAACGGTTTCAGTAATCCTCACTACTTGGGTCCAGATGTGAGGAACTTGAACCTGGCTCTGACCCCCGACTCGGGAGTTGCGCCTGGAGACATAGAGCTGCAAGACATGAACGTCGGTCGGCGGGCGGCCAGGCCCGGGGACACGCCACTGTACCTGCTCATCGTGGGCGGGAAGGAAACCCCGCACGTGGCCTTGATGCAGAGCCCTCTGTCTATGTGGGCTTATAGACTTCACTAA
Protein
MWSLVDSGGADGKSAPCSRGKHSATLLGGFVYVLGGRGSGGSVPLRDFWRYSLTTGNWEKLDCRGEHPPSLQEHTATAHGHRIYVFGGEAGALTNETPLWIYDTETQIWRKLPGQVNYTSSVRRRARDPRGAVSGPRGRRGHTAHALKDCLLIYGGYQDLKGSTNELWAFHYESESWHQVRTWNSGPARHRHAAALHDERLYVHGGQCDLRDCADLWFYDTMSRVWREVRARAGPAPRSGHAAARARAHLYIFGGDSHSHPHNDLWRFHFATETWEHIPNTVKWPSPRTDTCAILLPRRPPRAPPRSPPLAHPTLARPPPPPGNILREISKLSAFHIRRAARCSYTALAGNKDSTESLVPSESTLPKSQSAFHMERRGQCSPREGDVPDLTRDPLSVPDFADMVTTPTRTTKLVYLDSEDEANGKRDVTVKMDSETTVVMPKSASVRFTKTVTITDEEDTEMSTSDYASAEKVNRISGNGFSNPHYLGPDVRNLNLALTPDSGVAPGDIELQDMNVGRRAARPGDTPLYLLIVGGKETPHVALMQSPLSMWAYRLH
Summary
Uniprot
A0A2H1VK29
A0A2W1BFR7
A0A212FEM8
A0A194PQT4
A0A023F292
A0A0A9ZDW6
+ More
A0A146LGW2 A0A0L7R6V4 A0A0N0U7S5 A0A158NFK4 A0A088ASR5 A0A195D718 E2B0W6 A0A195DP75 F4X1B3 A0A3L8DC03 A0A154PF52 A0A0C9RWE8 A0A195BDQ1 A0A2A3E9K9 E9ITH8 A0A2R7WER2 N6UC05 A0A232FCH4 K7JD27 U4TY43 A0A151WNT5 A0A151JSX7 A0A026WWT8 E2C136
A0A146LGW2 A0A0L7R6V4 A0A0N0U7S5 A0A158NFK4 A0A088ASR5 A0A195D718 E2B0W6 A0A195DP75 F4X1B3 A0A3L8DC03 A0A154PF52 A0A0C9RWE8 A0A195BDQ1 A0A2A3E9K9 E9ITH8 A0A2R7WER2 N6UC05 A0A232FCH4 K7JD27 U4TY43 A0A151WNT5 A0A151JSX7 A0A026WWT8 E2C136
Pubmed
EMBL
ODYU01002990
SOQ41183.1
KZ150126
PZC73111.1
AGBW02008930
OWR52170.1
+ More
KQ459595 KPI95811.1 GBBI01003628 JAC15084.1 GBHO01000132 GBHO01000131 GBHO01000130 GBHO01000129 GBHO01000128 GBHO01000127 JAG43472.1 JAG43473.1 JAG43474.1 JAG43475.1 JAG43476.1 JAG43477.1 GDHC01012442 JAQ06187.1 KQ414646 KOC66578.1 KQ435700 KOX80451.1 ADTU01014559 ADTU01014560 ADTU01014561 KQ976750 KYN08685.1 GL444701 EFN60669.1 KQ980662 KYN14673.1 GL888528 EGI59785.1 QOIP01000010 RLU17884.1 KQ434890 KZC10479.1 GBYB01013289 GBYB01013290 JAG83056.1 JAG83057.1 KQ976514 KYM82310.1 KZ288334 PBC27879.1 GL765555 EFZ16128.1 KK854721 PTY18154.1 APGK01029532 APGK01029533 KB740735 ENN79185.1 NNAY01000490 OXU28027.1 KB631642 ERL84908.1 KQ982893 KYQ49562.1 KQ982011 KYN30672.1 KK107079 EZA60171.1 GL451853 EFN78357.1
KQ459595 KPI95811.1 GBBI01003628 JAC15084.1 GBHO01000132 GBHO01000131 GBHO01000130 GBHO01000129 GBHO01000128 GBHO01000127 JAG43472.1 JAG43473.1 JAG43474.1 JAG43475.1 JAG43476.1 JAG43477.1 GDHC01012442 JAQ06187.1 KQ414646 KOC66578.1 KQ435700 KOX80451.1 ADTU01014559 ADTU01014560 ADTU01014561 KQ976750 KYN08685.1 GL444701 EFN60669.1 KQ980662 KYN14673.1 GL888528 EGI59785.1 QOIP01000010 RLU17884.1 KQ434890 KZC10479.1 GBYB01013289 GBYB01013290 JAG83056.1 JAG83057.1 KQ976514 KYM82310.1 KZ288334 PBC27879.1 GL765555 EFZ16128.1 KK854721 PTY18154.1 APGK01029532 APGK01029533 KB740735 ENN79185.1 NNAY01000490 OXU28027.1 KB631642 ERL84908.1 KQ982893 KYQ49562.1 KQ982011 KYN30672.1 KK107079 EZA60171.1 GL451853 EFN78357.1
Proteomes
Interpro
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1VK29
A0A2W1BFR7
A0A212FEM8
A0A194PQT4
A0A023F292
A0A0A9ZDW6
+ More
A0A146LGW2 A0A0L7R6V4 A0A0N0U7S5 A0A158NFK4 A0A088ASR5 A0A195D718 E2B0W6 A0A195DP75 F4X1B3 A0A3L8DC03 A0A154PF52 A0A0C9RWE8 A0A195BDQ1 A0A2A3E9K9 E9ITH8 A0A2R7WER2 N6UC05 A0A232FCH4 K7JD27 U4TY43 A0A151WNT5 A0A151JSX7 A0A026WWT8 E2C136
A0A146LGW2 A0A0L7R6V4 A0A0N0U7S5 A0A158NFK4 A0A088ASR5 A0A195D718 E2B0W6 A0A195DP75 F4X1B3 A0A3L8DC03 A0A154PF52 A0A0C9RWE8 A0A195BDQ1 A0A2A3E9K9 E9ITH8 A0A2R7WER2 N6UC05 A0A232FCH4 K7JD27 U4TY43 A0A151WNT5 A0A151JSX7 A0A026WWT8 E2C136
PDB
6DO5
E-value=6.28268e-07,
Score=129
Ontologies
GO
Topology
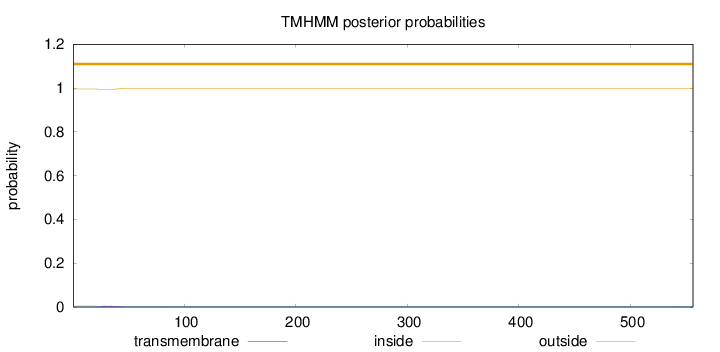
Length:
556
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1003
Exp number, first 60 AAs:
0.0961
Total prob of N-in:
0.00476
outside
1 - 556
Population Genetic Test Statistics
Pi
14.977429
Theta
14.366173
Tajima's D
0.395822
CLR
11.657522
CSRT
0.498925053747313
Interpretation
Uncertain
