Gene
KWMTBOMO04824
Pre Gene Modal
BGIBMGA009827
Annotation
PREDICTED:_uncharacterized_protein_LOC106142321_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.876 PlasmaMembrane Reliability : 1.203
Sequence
CDS
ATGGGCTTTAGTAACCACTCGGACGGATCTCAAGTCGGCACAATGGTTGGCACTAACAATTTGCGAAGCAATCTAAGCGACGTGTTCAGTGAAAAGCAATTAAGTCAAATAGTCAAATCATCAGTGGGCGAGGGTGGGAAAGTGATTGATGGGTACGTCAAACCTGTCGCTGATGGCATAGCAGGATTCCTCGGTGATCATTTTAAAGTGACTTTAGAAGTACAAGTGAGTGGACAAGTTACATGTTTGCATTTATTCGTGAAACGATTGCCTGTTAACAATAAACCGAAGGCTGAGTTCATCGACAGTCAGCACTTTTTTAGAAGAGAGCGGTTAATTTTTGAACTTTTTGAAGAATTTGAGGACGTCGGAGATCCGAATCCATGGAACGTCAAAGCATACCTATACTCCGACAGCATGCTGGTCCTGCCCGACTTGGACATCCAGGGCTATCGTTCTCGGCACCACCTGGATACCTTCGACCTACCACACGCGACAGTCACCATTACGTCCATCGCTCGTTTCCACGCAACCTTCGCTAATTACGAGACTAAGAATATCCTCAACGGAAGAAGTGAATACAATTTTTACAAACAACACGAACACTTATTAAACGAACCCACTTTTAAGGACTGCCCTTGGCTTCGTGCGGCCGCAAAATGTATTTCAGACATCCTAAACACTTTCTCCGCAAAATACGTTGGCTTGCCAGAATTAGAAAATAGTATTGTAAAATTGTTTATAGAAGCTTGTGATAGTTTGACTGTAGTTAAAGATGGATTCAATGTATTGATACACAAAGACTTATGGGCTAATAACATCATGTTTAAATACGAGGACTGCAATCCTAATAATGCTGTTATCATAGATTACCAATGTATAAGATACTCGGCACCGACGTTTGATGTCATGACCTTTCTGTACCTGACGACATCGAAGAGTTTCCGGCAGCAGCACGAAACAGGAATCTTCGAGCACTACTATACAATTTTTTCCAAGCATCTAAATGAAAATACCAAATTGAGATTGAAAAGAATCGGCTACAACTGGGAGGAATTTTTGAAGAGTTGCGAAAACTCTAGAATGTTTGGTATTTTTCAAGCTTTGGGTATATGTCCTTATATCTTATTAGATCCGAAGACTGCTGCAGCTACTTTTGACGATCCGGAAACCTTTCAGAAGCATAACTTCGAAGATAGGTCTGCCCCTTTAATACGCTACGCGAGGACCTGCAATGTATACCGGGAGAGACTGGTAGAATTGTCTGAGGAATTCGTGGAAAGATATTTATTGAAAAAATAA
Protein
MGFSNHSDGSQVGTMVGTNNLRSNLSDVFSEKQLSQIVKSSVGEGGKVIDGYVKPVADGIAGFLGDHFKVTLEVQVSGQVTCLHLFVKRLPVNNKPKAEFIDSQHFFRRERLIFELFEEFEDVGDPNPWNVKAYLYSDSMLVLPDLDIQGYRSRHHLDTFDLPHATVTITSIARFHATFANYETKNILNGRSEYNFYKQHEHLLNEPTFKDCPWLRAAAKCISDILNTFSAKYVGLPELENSIVKLFIEACDSLTVVKDGFNVLIHKDLWANNIMFKYEDCNPNNAVIIDYQCIRYSAPTFDVMTFLYLTTSKSFRQQHETGIFEHYYTIFSKHLNENTKLRLKRIGYNWEEFLKSCENSRMFGIFQALGICPYILLDPKTAAATFDDPETFQKHNFEDRSAPLIRYARTCNVYRERLVELSEEFVERYLLKK
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
H9JJX6
A0A2W1BE00
A0A2A4JSU1
A0A2H1V3K8
A0A0L7L3I9
A0A194RD92
+ More
A0A194PQS6 A0A1E1WGR0 A0A212FEP4 A0A194PM76 A0A194QSH4 A0A194PN60 A0A2H1VY68 A0A194QXY3 A0A310SGN2 A0A0L7LBN0 A0A0L7R7K3 D6WK24 A0A088AQ18 A0A2A3EI69 B0X965 K7J3E6 E2AYB7 E2AYB9 A0A0L7L3M8 K7J3E7 H9J767 H9J766 A0A154PDS1 A0A212EU23 A0A0J7KMP1 A0A1S4FE87 A0A1S4FE70 A0A195E6F3 A0A195EUG8 U4U5Y9 Q175M8 N6UGU5 A0A026WP10 A0A3L8DXD2 E2AYC0 A0A182G3W0 E9ISQ3 A0A0P6J630 K7J940 A0A0L7QQ26 A0A182GQL4 F4WPC3 A0A0Q9WRB1 E2BN44 A0A158NQX7 A0A195B0G2 K7IPN3 E9JBH4 E2BDR5 A0A151WMN2 A0A1W4UJ13 A0A212EU10 A0A182WT65 A0A182HV60 A0A182GQL3 B4LUL7 A0A1J1HSN4 A0A088A736 A0A2A3E710 V9IFX7 K7IPV1 A0A182YGJ3 Q71DB6 E1ZZJ3 A0A182W7V2 A0A0P8XHW5 A0A3L8E2M6 B4JE98 A0A232FKC4 A0A1W7R8B2 A0A182NEB7 A0A0T6BHU0 A0A232EK39 A0A1S4HCD0 A0A0M9A434 D6WK27 Q9VIG3 A0A182VSC8 Q4V4W6 B4IFK0 D2A2D6 A0A069DU06 A0A2P8YLX0 A0A088AES4 B4Q3Y1 U5EDE8 A0A2A3ECT0 A0A0J7KTS7 K7J679 A0A182PF65 F4WR92 T1HUP9 A0A1Q3FRJ8
A0A194PQS6 A0A1E1WGR0 A0A212FEP4 A0A194PM76 A0A194QSH4 A0A194PN60 A0A2H1VY68 A0A194QXY3 A0A310SGN2 A0A0L7LBN0 A0A0L7R7K3 D6WK24 A0A088AQ18 A0A2A3EI69 B0X965 K7J3E6 E2AYB7 E2AYB9 A0A0L7L3M8 K7J3E7 H9J767 H9J766 A0A154PDS1 A0A212EU23 A0A0J7KMP1 A0A1S4FE87 A0A1S4FE70 A0A195E6F3 A0A195EUG8 U4U5Y9 Q175M8 N6UGU5 A0A026WP10 A0A3L8DXD2 E2AYC0 A0A182G3W0 E9ISQ3 A0A0P6J630 K7J940 A0A0L7QQ26 A0A182GQL4 F4WPC3 A0A0Q9WRB1 E2BN44 A0A158NQX7 A0A195B0G2 K7IPN3 E9JBH4 E2BDR5 A0A151WMN2 A0A1W4UJ13 A0A212EU10 A0A182WT65 A0A182HV60 A0A182GQL3 B4LUL7 A0A1J1HSN4 A0A088A736 A0A2A3E710 V9IFX7 K7IPV1 A0A182YGJ3 Q71DB6 E1ZZJ3 A0A182W7V2 A0A0P8XHW5 A0A3L8E2M6 B4JE98 A0A232FKC4 A0A1W7R8B2 A0A182NEB7 A0A0T6BHU0 A0A232EK39 A0A1S4HCD0 A0A0M9A434 D6WK27 Q9VIG3 A0A182VSC8 Q4V4W6 B4IFK0 D2A2D6 A0A069DU06 A0A2P8YLX0 A0A088AES4 B4Q3Y1 U5EDE8 A0A2A3ECT0 A0A0J7KTS7 K7J679 A0A182PF65 F4WR92 T1HUP9 A0A1Q3FRJ8
Pubmed
19121390
28756777
26227816
26354079
22118469
18362917
+ More
19820115 20075255 20798317 23537049 17510324 24508170 30249741 26483478 21282665 26999592 21719571 17994087 21347285 25244985 14525923 17550304 28648823 12364791 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 29403074 22936249
19820115 20075255 20798317 23537049 17510324 24508170 30249741 26483478 21282665 26999592 21719571 17994087 21347285 25244985 14525923 17550304 28648823 12364791 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 29403074 22936249
EMBL
BABH01026750
KZ150126
PZC73108.1
NWSH01000742
PCG74462.1
ODYU01000324
+ More
SOQ34864.1 JTDY01003245 KOB69886.1 KQ460367 KPJ15439.1 KQ459595 KPI95801.1 GDQN01005003 JAT86051.1 AGBW02008930 OWR52167.1 KQ459599 KPI94427.1 KQ461154 KPJ08437.1 KPI94428.1 ODYU01005000 SOQ45442.1 KPJ08436.1 KQ761346 OAD57837.1 JTDY01001890 KOB72621.1 KQ414639 KOC66818.1 KQ971342 EFA03937.1 KZ288232 PBC31485.1 DS232520 EDS42971.1 GL443864 EFN61576.1 EFN61578.1 JTDY01003170 KOB70020.1 BABH01013774 KQ434869 KZC09408.1 AGBW02012491 OWR44993.1 LBMM01005433 KMQ91529.1 KQ979608 KYN20424.1 KQ981965 KYN31801.1 KB632064 ERL88482.1 CH477398 EAT41774.1 APGK01035612 KB740928 ENN77872.1 KK107139 EZA57755.1 QOIP01000003 RLU25036.1 EFN61579.1 JXUM01142400 KQ569416 KXJ68636.1 GL765434 EFZ16323.1 GDUN01000271 JAN95648.1 AAZX01018058 KQ414807 KOC60591.1 JXUM01080854 KQ563212 KXJ74284.1 GL888243 EGI64018.1 CH963920 KRF98755.1 GL449382 EFN82948.1 ADTU01023697 KQ976691 KYM77968.1 GL771089 EFZ09829.1 GL447689 EFN86121.1 KQ982944 KYQ48965.1 OWR44992.1 APCN01002494 KXJ74283.1 CH940649 EDW64203.1 CVRI01000020 CRK90554.1 KZ288353 PBC27284.1 JR045443 AEY59980.1 AF531953 CM000158 AAQ09852.1 EDW89971.1 GL435377 EFN73398.1 CH902624 KPU74451.1 QOIP01000001 RLU26449.1 CH916368 EDW03618.1 NNAY01000087 OXU31125.1 GEHC01000254 JAV47391.1 LJIG01000093 KRT86847.1 NNAY01003909 OXU18671.1 AAAB01008964 KQ435762 KOX75449.1 EFA03938.1 AE014134 BT128869 AAF53956.1 AEN86879.1 BT022890 AAY55306.1 CH480833 EDW46422.1 KQ971338 EFA02038.1 GBGD01001712 JAC87177.1 PYGN01000502 PSN45249.1 CM000361 CM002910 EDX05697.1 KMY91273.1 GANO01004792 JAB55079.1 KZ288301 PBC28831.1 LBMM01003342 KMQ93639.1 GL888284 EGI63309.1 ACPB03005212 GFDL01004814 JAV30231.1
SOQ34864.1 JTDY01003245 KOB69886.1 KQ460367 KPJ15439.1 KQ459595 KPI95801.1 GDQN01005003 JAT86051.1 AGBW02008930 OWR52167.1 KQ459599 KPI94427.1 KQ461154 KPJ08437.1 KPI94428.1 ODYU01005000 SOQ45442.1 KPJ08436.1 KQ761346 OAD57837.1 JTDY01001890 KOB72621.1 KQ414639 KOC66818.1 KQ971342 EFA03937.1 KZ288232 PBC31485.1 DS232520 EDS42971.1 GL443864 EFN61576.1 EFN61578.1 JTDY01003170 KOB70020.1 BABH01013774 KQ434869 KZC09408.1 AGBW02012491 OWR44993.1 LBMM01005433 KMQ91529.1 KQ979608 KYN20424.1 KQ981965 KYN31801.1 KB632064 ERL88482.1 CH477398 EAT41774.1 APGK01035612 KB740928 ENN77872.1 KK107139 EZA57755.1 QOIP01000003 RLU25036.1 EFN61579.1 JXUM01142400 KQ569416 KXJ68636.1 GL765434 EFZ16323.1 GDUN01000271 JAN95648.1 AAZX01018058 KQ414807 KOC60591.1 JXUM01080854 KQ563212 KXJ74284.1 GL888243 EGI64018.1 CH963920 KRF98755.1 GL449382 EFN82948.1 ADTU01023697 KQ976691 KYM77968.1 GL771089 EFZ09829.1 GL447689 EFN86121.1 KQ982944 KYQ48965.1 OWR44992.1 APCN01002494 KXJ74283.1 CH940649 EDW64203.1 CVRI01000020 CRK90554.1 KZ288353 PBC27284.1 JR045443 AEY59980.1 AF531953 CM000158 AAQ09852.1 EDW89971.1 GL435377 EFN73398.1 CH902624 KPU74451.1 QOIP01000001 RLU26449.1 CH916368 EDW03618.1 NNAY01000087 OXU31125.1 GEHC01000254 JAV47391.1 LJIG01000093 KRT86847.1 NNAY01003909 OXU18671.1 AAAB01008964 KQ435762 KOX75449.1 EFA03938.1 AE014134 BT128869 AAF53956.1 AEN86879.1 BT022890 AAY55306.1 CH480833 EDW46422.1 KQ971338 EFA02038.1 GBGD01001712 JAC87177.1 PYGN01000502 PSN45249.1 CM000361 CM002910 EDX05697.1 KMY91273.1 GANO01004792 JAB55079.1 KZ288301 PBC28831.1 LBMM01003342 KMQ93639.1 GL888284 EGI63309.1 ACPB03005212 GFDL01004814 JAV30231.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000053825 UP000007266 UP000005203 UP000242457 UP000002320 UP000002358 UP000000311 UP000076502 UP000036403 UP000078492 UP000078541 UP000030742 UP000008820 UP000019118 UP000053097 UP000279307 UP000069940 UP000249989 UP000007755 UP000007798 UP000008237 UP000005205 UP000078540 UP000075809 UP000192221 UP000076407 UP000075840 UP000008792 UP000183832 UP000076408 UP000002282 UP000075920 UP000007801 UP000001070 UP000215335 UP000075884 UP000053105 UP000000803 UP000001292 UP000245037 UP000000304 UP000075885 UP000015103
UP000053825 UP000007266 UP000005203 UP000242457 UP000002320 UP000002358 UP000000311 UP000076502 UP000036403 UP000078492 UP000078541 UP000030742 UP000008820 UP000019118 UP000053097 UP000279307 UP000069940 UP000249989 UP000007755 UP000007798 UP000008237 UP000005205 UP000078540 UP000075809 UP000192221 UP000076407 UP000075840 UP000008792 UP000183832 UP000076408 UP000002282 UP000075920 UP000007801 UP000001070 UP000215335 UP000075884 UP000053105 UP000000803 UP000001292 UP000245037 UP000000304 UP000075885 UP000015103
Interpro
Gene 3D
ProteinModelPortal
H9JJX6
A0A2W1BE00
A0A2A4JSU1
A0A2H1V3K8
A0A0L7L3I9
A0A194RD92
+ More
A0A194PQS6 A0A1E1WGR0 A0A212FEP4 A0A194PM76 A0A194QSH4 A0A194PN60 A0A2H1VY68 A0A194QXY3 A0A310SGN2 A0A0L7LBN0 A0A0L7R7K3 D6WK24 A0A088AQ18 A0A2A3EI69 B0X965 K7J3E6 E2AYB7 E2AYB9 A0A0L7L3M8 K7J3E7 H9J767 H9J766 A0A154PDS1 A0A212EU23 A0A0J7KMP1 A0A1S4FE87 A0A1S4FE70 A0A195E6F3 A0A195EUG8 U4U5Y9 Q175M8 N6UGU5 A0A026WP10 A0A3L8DXD2 E2AYC0 A0A182G3W0 E9ISQ3 A0A0P6J630 K7J940 A0A0L7QQ26 A0A182GQL4 F4WPC3 A0A0Q9WRB1 E2BN44 A0A158NQX7 A0A195B0G2 K7IPN3 E9JBH4 E2BDR5 A0A151WMN2 A0A1W4UJ13 A0A212EU10 A0A182WT65 A0A182HV60 A0A182GQL3 B4LUL7 A0A1J1HSN4 A0A088A736 A0A2A3E710 V9IFX7 K7IPV1 A0A182YGJ3 Q71DB6 E1ZZJ3 A0A182W7V2 A0A0P8XHW5 A0A3L8E2M6 B4JE98 A0A232FKC4 A0A1W7R8B2 A0A182NEB7 A0A0T6BHU0 A0A232EK39 A0A1S4HCD0 A0A0M9A434 D6WK27 Q9VIG3 A0A182VSC8 Q4V4W6 B4IFK0 D2A2D6 A0A069DU06 A0A2P8YLX0 A0A088AES4 B4Q3Y1 U5EDE8 A0A2A3ECT0 A0A0J7KTS7 K7J679 A0A182PF65 F4WR92 T1HUP9 A0A1Q3FRJ8
A0A194PQS6 A0A1E1WGR0 A0A212FEP4 A0A194PM76 A0A194QSH4 A0A194PN60 A0A2H1VY68 A0A194QXY3 A0A310SGN2 A0A0L7LBN0 A0A0L7R7K3 D6WK24 A0A088AQ18 A0A2A3EI69 B0X965 K7J3E6 E2AYB7 E2AYB9 A0A0L7L3M8 K7J3E7 H9J767 H9J766 A0A154PDS1 A0A212EU23 A0A0J7KMP1 A0A1S4FE87 A0A1S4FE70 A0A195E6F3 A0A195EUG8 U4U5Y9 Q175M8 N6UGU5 A0A026WP10 A0A3L8DXD2 E2AYC0 A0A182G3W0 E9ISQ3 A0A0P6J630 K7J940 A0A0L7QQ26 A0A182GQL4 F4WPC3 A0A0Q9WRB1 E2BN44 A0A158NQX7 A0A195B0G2 K7IPN3 E9JBH4 E2BDR5 A0A151WMN2 A0A1W4UJ13 A0A212EU10 A0A182WT65 A0A182HV60 A0A182GQL3 B4LUL7 A0A1J1HSN4 A0A088A736 A0A2A3E710 V9IFX7 K7IPV1 A0A182YGJ3 Q71DB6 E1ZZJ3 A0A182W7V2 A0A0P8XHW5 A0A3L8E2M6 B4JE98 A0A232FKC4 A0A1W7R8B2 A0A182NEB7 A0A0T6BHU0 A0A232EK39 A0A1S4HCD0 A0A0M9A434 D6WK27 Q9VIG3 A0A182VSC8 Q4V4W6 B4IFK0 D2A2D6 A0A069DU06 A0A2P8YLX0 A0A088AES4 B4Q3Y1 U5EDE8 A0A2A3ECT0 A0A0J7KTS7 K7J679 A0A182PF65 F4WR92 T1HUP9 A0A1Q3FRJ8
Ontologies
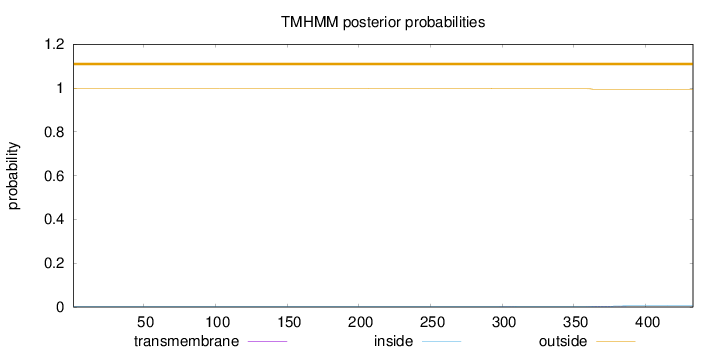
Topology
Length:
433
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05444
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00278
outside
1 - 433
Population Genetic Test Statistics
Pi
8.365778
Theta
10.97442
Tajima's D
-0.797896
CLR
1.196245
CSRT
0.176291185440728
Interpretation
Uncertain
