Gene
KWMTBOMO04821 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009830
Annotation
eukaryotic_translation_initiation_factor_6_[Bombyx_mori]
Full name
Eukaryotic translation initiation factor 6
Location in the cell
Cytoplasmic Reliability : 1.716
Sequence
CDS
ATGGCGGTTCGCGTTCAATTTGAAAATAATAATGAAGTGGGAGTGTTCAGTAAATTAACAAACACATACTGCCTTGTCGCGATTGGAGGTTCGGAGAATTTTTACAGCGTGTTCGAAGCTGAATTAGCGGAGACCATCCCAGTGATACATGCGAGTGTTGGAGGCTGCCGAATCATCGGTAGAATGACCGTAGGGAACAAGAACGGTCTCTTAGTTCCATCTTCGACCACGGACACAGAATTACAACACATTAGAAACAGTCTGCCGGAGAACGTGAAATTACAACGCGTGGAGGAACGGCTTAGTGCCCTCGGTAACGTCATAGCCTGTAACGACTATGTAGCACTTGTACATCCTGACTTAGACAAAGACACAGAAGAAATATTAGCCGACACATTAAACGTGGAAGTATTCAGACAAACAATAGCGGGAAACGTACTTGTTGGTTCTTACACTGCATTGAGCAACCGAGGAGGGCTGGTACACCCGAAGACAAATATACAGGACCAGGATGAACTCTCTTCGCTACTCCAAGTGCCCTTAGTTGCCGGAACAGTTAACAGAGGCAGTGATGTCGTAGCTGCTGGAATGGTGGTCAACGATTGGTGTGCATTCTGCGGACTGGACACAACGTCAACTGAGATTTCTGTTATTGAGAGTGTTTTCAAATTAAATGATGCTAAACCGACTGCAATCACATCTACAATGAGAGCATCGCTCATTGACAGTATGTCATAA
Protein
MAVRVQFENNNEVGVFSKLTNTYCLVAIGGSENFYSVFEAELAETIPVIHASVGGCRIIGRMTVGNKNGLLVPSSTTDTELQHIRNSLPENVKLQRVEERLSALGNVIACNDYVALVHPDLDKDTEEILADTLNVEVFRQTIAGNVLVGSYTALSNRGGLVHPKTNIQDQDELSSLLQVPLVAGTVNRGSDVVAAGMVVNDWCAFCGLDTTSTEISVIESVFKLNDAKPTAITSTMRASLIDSMS
Summary
Description
Binds to the 60S ribosomal subunit and prevents its association with the 40S ribosomal subunit to form the 80S initiation complex in the cytoplasm. May also be involved in ribosome biogenesis.
Subunit
Monomer. Associates with the 60S ribosomal subunit.
Similarity
Belongs to the eIF-6 family.
Keywords
Cytoplasm
Initiation factor
Nucleus
Protein biosynthesis
Ribosome biogenesis
Feature
chain Eukaryotic translation initiation factor 6
Uniprot
Q1HQ59
A0A0L7LH40
A0S6A4
A0A212FEJ5
A0A1E1WBJ3
A0A2W1BHC2
+ More
A0A2A4JS04 A0A194PSS3 A0A3S2NZ78 A0A194RD87 S4P457 A0A088A2P5 E0VVM1 A0A0V0G988 A0A2A3EH32 A0A1S4EIA4 A0A067QMJ9 K7IPC1 A0A0P4W221 R4G4M6 A0A224XV59 A0A023F9D2 E2BSI5 R4WJE1 A0A2R7W8G1 A0A023GGS0 A0A158NE26 A0A1B6DEM1 G3MMH3 A0A1B6G0I8 A0A1B6IX34 A0A023FXX0 A0A1E1XIB0 A0A131XI39 A0A224Z9Z6 A0A131Z3M0 L7M4J9 A0A023FGL5 A0A0C9S1Z6 A0A0K8TRQ2 A0A1B6K9R5 F4W519 A0A0A9YU28 A0A2P6LBR9 A0A195BQL5 A0A195F545 A0A151XED4 A0A151IF09 A0A0L7RB36 A0A087U4V8 A0A195D9U9 A0A2R5LF86 A0A131XUQ6 A0A026WZX9 A0A1Z5LHA1 H2LCJ9 A0A3P8WUI6 A0A3B5BCR8 E2AQP0 A0A3B4DSN8 A0A1Y1L5Y5 I5APZ0 A0A3Q3LQP7 A0A3B3BAX9 A0A0Q9XAB8 A0A0P8Y1K9 A0A3Q2PSI8 A0A1L8EDA3 A0A3P9JY68 E9J331 B4K0F6 A0A3Q0RLG6 A0A1A8EY25 A0A0M3QUM4 A0A1A7ZLU5 A0A3Q1J9L5 A0A3P8U4V6 A0A1I8MIU5 A0A1A8JQ36 A0A1A8CW35 A0A3B4XE96 A0A3B4V057 A0A1L8ES41 A0A2U9BEW4 A0A3B3QPR3 A0A3Q1CBD9 A0A293N4Q0 A0A3B1JW97 A0A1A8SIU3 A0A1A8MN39 A0A1A7X9K4 V5HK20 A0A3S2N4J7 Q6GR45 H2YS09 B4LIY3 A0A1I8PD35 A0A2I4BSR8 C1BL53 W5MM18
A0A2A4JS04 A0A194PSS3 A0A3S2NZ78 A0A194RD87 S4P457 A0A088A2P5 E0VVM1 A0A0V0G988 A0A2A3EH32 A0A1S4EIA4 A0A067QMJ9 K7IPC1 A0A0P4W221 R4G4M6 A0A224XV59 A0A023F9D2 E2BSI5 R4WJE1 A0A2R7W8G1 A0A023GGS0 A0A158NE26 A0A1B6DEM1 G3MMH3 A0A1B6G0I8 A0A1B6IX34 A0A023FXX0 A0A1E1XIB0 A0A131XI39 A0A224Z9Z6 A0A131Z3M0 L7M4J9 A0A023FGL5 A0A0C9S1Z6 A0A0K8TRQ2 A0A1B6K9R5 F4W519 A0A0A9YU28 A0A2P6LBR9 A0A195BQL5 A0A195F545 A0A151XED4 A0A151IF09 A0A0L7RB36 A0A087U4V8 A0A195D9U9 A0A2R5LF86 A0A131XUQ6 A0A026WZX9 A0A1Z5LHA1 H2LCJ9 A0A3P8WUI6 A0A3B5BCR8 E2AQP0 A0A3B4DSN8 A0A1Y1L5Y5 I5APZ0 A0A3Q3LQP7 A0A3B3BAX9 A0A0Q9XAB8 A0A0P8Y1K9 A0A3Q2PSI8 A0A1L8EDA3 A0A3P9JY68 E9J331 B4K0F6 A0A3Q0RLG6 A0A1A8EY25 A0A0M3QUM4 A0A1A7ZLU5 A0A3Q1J9L5 A0A3P8U4V6 A0A1I8MIU5 A0A1A8JQ36 A0A1A8CW35 A0A3B4XE96 A0A3B4V057 A0A1L8ES41 A0A2U9BEW4 A0A3B3QPR3 A0A3Q1CBD9 A0A293N4Q0 A0A3B1JW97 A0A1A8SIU3 A0A1A8MN39 A0A1A7X9K4 V5HK20 A0A3S2N4J7 Q6GR45 H2YS09 B4LIY3 A0A1I8PD35 A0A2I4BSR8 C1BL53 W5MM18
Pubmed
19121390
26227816
22118469
28756777
26354079
23622113
+ More
20566863 24845553 20075255 27129103 25474469 20798317 23691247 21347285 22216098 28503490 28049606 28797301 26830274 25576852 26131772 26369729 21719571 25401762 26823975 24508170 30249741 28528879 17554307 24487278 28004739 15632085 17994087 29451363 21282665 25315136 27762356 29240929 25329095 25765539 16425228
20566863 24845553 20075255 27129103 25474469 20798317 23691247 21347285 22216098 28503490 28049606 28797301 26830274 25576852 26131772 26369729 21719571 25401762 26823975 24508170 30249741 28528879 17554307 24487278 28004739 15632085 17994087 29451363 21282665 25315136 27762356 29240929 25329095 25765539 16425228
EMBL
BABH01026740
DQ443193
ABF51282.1
JTDY01001166
KOB74674.1
DQ289585
+ More
ODYU01006120 ABB92838.1 SOQ47692.1 AGBW02008930 OWR52161.1 GDQN01006739 JAT84315.1 KZ150126 PZC73105.1 NWSH01000742 PCG74468.1 KQ459595 KPI95809.1 RSAL01000018 RVE52872.1 KQ460367 KPJ15434.1 GAIX01006144 JAA86416.1 AAZO01005657 DS235812 EEB17427.1 GECL01001473 JAP04651.1 KZ288252 PBC31030.1 KK853149 KDR10668.1 GDKW01000929 JAI55666.1 GAHY01000641 JAA76869.1 GFTR01004044 JAW12382.1 GBBI01000722 JAC17990.1 GL450240 EFN81318.1 AK417640 BAN20855.1 KK854459 PTY16017.1 GBBM01002266 JAC33152.1 ADTU01012616 ADTU01012617 GEDC01013253 JAS24045.1 JO843074 AEO34691.1 GECZ01013811 JAS55958.1 GECU01016210 JAS91496.1 GBBL01001752 JAC25568.1 GFAC01000362 JAT98826.1 GEFH01002773 JAP65808.1 GFPF01012737 MAA23883.1 GEDV01002650 JAP85907.1 GACK01005783 JAA59251.1 GBBK01003456 JAC21026.1 GBZX01001801 JAG90939.1 GDAI01000561 JAI17042.1 GEBQ01031791 GEBQ01014878 JAT08186.1 JAT25099.1 GL887596 EGI70715.1 GBHO01008503 GBRD01010348 GDHC01018468 JAG35101.1 JAG55476.1 JAQ00161.1 MWRG01000475 PRD36025.1 KQ976424 KYM88256.1 KQ981820 KYN35194.1 KQ982254 KYQ58731.1 KQ977830 KYM99407.1 KQ414617 KOC68132.1 KK118176 KFM72397.1 KQ981082 KYN09658.1 GGLE01004067 MBY08193.1 GEFM01006495 JAP69301.1 KK107054 QOIP01000004 EZA61595.1 RLU23485.1 GFJQ02000432 JAW06538.1 GL441804 EFN64229.1 GEZM01066587 JAV67780.1 CM000071 EIM53025.1 CH933808 KRG05440.1 CH902619 KPU75685.1 GFDG01002167 JAV16632.1 GL768055 EFZ12818.1 CH916444 EDV95481.1 HAEB01004227 HAEC01013355 SBQ50754.1 CP012524 ALC40883.1 HADY01005327 HAEJ01006632 SBP43812.1 HAED01018135 HAEE01002203 SBR22223.1 HADZ01020071 HAEA01003468 SBP84012.1 CM004482 OCT62164.1 CP026248 AWP02349.1 GFWV01022662 MAA47389.1 HAEH01004523 HAEI01015673 SBS18142.1 HAEF01016685 HAEG01005974 SBR57844.1 HADW01013392 HADX01003783 SBP14792.1 GANP01006539 JAB77929.1 CM012441 RVE73357.1 AY383654 BC071088 CH940648 EDW60433.1 BT075332 ACO09756.1 AHAT01024190
ODYU01006120 ABB92838.1 SOQ47692.1 AGBW02008930 OWR52161.1 GDQN01006739 JAT84315.1 KZ150126 PZC73105.1 NWSH01000742 PCG74468.1 KQ459595 KPI95809.1 RSAL01000018 RVE52872.1 KQ460367 KPJ15434.1 GAIX01006144 JAA86416.1 AAZO01005657 DS235812 EEB17427.1 GECL01001473 JAP04651.1 KZ288252 PBC31030.1 KK853149 KDR10668.1 GDKW01000929 JAI55666.1 GAHY01000641 JAA76869.1 GFTR01004044 JAW12382.1 GBBI01000722 JAC17990.1 GL450240 EFN81318.1 AK417640 BAN20855.1 KK854459 PTY16017.1 GBBM01002266 JAC33152.1 ADTU01012616 ADTU01012617 GEDC01013253 JAS24045.1 JO843074 AEO34691.1 GECZ01013811 JAS55958.1 GECU01016210 JAS91496.1 GBBL01001752 JAC25568.1 GFAC01000362 JAT98826.1 GEFH01002773 JAP65808.1 GFPF01012737 MAA23883.1 GEDV01002650 JAP85907.1 GACK01005783 JAA59251.1 GBBK01003456 JAC21026.1 GBZX01001801 JAG90939.1 GDAI01000561 JAI17042.1 GEBQ01031791 GEBQ01014878 JAT08186.1 JAT25099.1 GL887596 EGI70715.1 GBHO01008503 GBRD01010348 GDHC01018468 JAG35101.1 JAG55476.1 JAQ00161.1 MWRG01000475 PRD36025.1 KQ976424 KYM88256.1 KQ981820 KYN35194.1 KQ982254 KYQ58731.1 KQ977830 KYM99407.1 KQ414617 KOC68132.1 KK118176 KFM72397.1 KQ981082 KYN09658.1 GGLE01004067 MBY08193.1 GEFM01006495 JAP69301.1 KK107054 QOIP01000004 EZA61595.1 RLU23485.1 GFJQ02000432 JAW06538.1 GL441804 EFN64229.1 GEZM01066587 JAV67780.1 CM000071 EIM53025.1 CH933808 KRG05440.1 CH902619 KPU75685.1 GFDG01002167 JAV16632.1 GL768055 EFZ12818.1 CH916444 EDV95481.1 HAEB01004227 HAEC01013355 SBQ50754.1 CP012524 ALC40883.1 HADY01005327 HAEJ01006632 SBP43812.1 HAED01018135 HAEE01002203 SBR22223.1 HADZ01020071 HAEA01003468 SBP84012.1 CM004482 OCT62164.1 CP026248 AWP02349.1 GFWV01022662 MAA47389.1 HAEH01004523 HAEI01015673 SBS18142.1 HAEF01016685 HAEG01005974 SBR57844.1 HADW01013392 HADX01003783 SBP14792.1 GANP01006539 JAB77929.1 CM012441 RVE73357.1 AY383654 BC071088 CH940648 EDW60433.1 BT075332 ACO09756.1 AHAT01024190
Proteomes
UP000005204
UP000037510
UP000007151
UP000218220
UP000053268
UP000283053
+ More
UP000053240 UP000005203 UP000009046 UP000242457 UP000079169 UP000027135 UP000002358 UP000008237 UP000005205 UP000007755 UP000078540 UP000078541 UP000075809 UP000078542 UP000053825 UP000054359 UP000078492 UP000053097 UP000279307 UP000001038 UP000265120 UP000261400 UP000000311 UP000261440 UP000001819 UP000261640 UP000261560 UP000009192 UP000007801 UP000265000 UP000265180 UP000001070 UP000261340 UP000092553 UP000265040 UP000265080 UP000095301 UP000261360 UP000261420 UP000186698 UP000246464 UP000261540 UP000257160 UP000018467 UP000007875 UP000008792 UP000095300 UP000192220 UP000018468
UP000053240 UP000005203 UP000009046 UP000242457 UP000079169 UP000027135 UP000002358 UP000008237 UP000005205 UP000007755 UP000078540 UP000078541 UP000075809 UP000078542 UP000053825 UP000054359 UP000078492 UP000053097 UP000279307 UP000001038 UP000265120 UP000261400 UP000000311 UP000261440 UP000001819 UP000261640 UP000261560 UP000009192 UP000007801 UP000265000 UP000265180 UP000001070 UP000261340 UP000092553 UP000265040 UP000265080 UP000095301 UP000261360 UP000261420 UP000186698 UP000246464 UP000261540 UP000257160 UP000018467 UP000007875 UP000008792 UP000095300 UP000192220 UP000018468
Pfam
PF01912 eIF-6
Interpro
IPR002769
eIF6
CDD
ProteinModelPortal
Q1HQ59
A0A0L7LH40
A0S6A4
A0A212FEJ5
A0A1E1WBJ3
A0A2W1BHC2
+ More
A0A2A4JS04 A0A194PSS3 A0A3S2NZ78 A0A194RD87 S4P457 A0A088A2P5 E0VVM1 A0A0V0G988 A0A2A3EH32 A0A1S4EIA4 A0A067QMJ9 K7IPC1 A0A0P4W221 R4G4M6 A0A224XV59 A0A023F9D2 E2BSI5 R4WJE1 A0A2R7W8G1 A0A023GGS0 A0A158NE26 A0A1B6DEM1 G3MMH3 A0A1B6G0I8 A0A1B6IX34 A0A023FXX0 A0A1E1XIB0 A0A131XI39 A0A224Z9Z6 A0A131Z3M0 L7M4J9 A0A023FGL5 A0A0C9S1Z6 A0A0K8TRQ2 A0A1B6K9R5 F4W519 A0A0A9YU28 A0A2P6LBR9 A0A195BQL5 A0A195F545 A0A151XED4 A0A151IF09 A0A0L7RB36 A0A087U4V8 A0A195D9U9 A0A2R5LF86 A0A131XUQ6 A0A026WZX9 A0A1Z5LHA1 H2LCJ9 A0A3P8WUI6 A0A3B5BCR8 E2AQP0 A0A3B4DSN8 A0A1Y1L5Y5 I5APZ0 A0A3Q3LQP7 A0A3B3BAX9 A0A0Q9XAB8 A0A0P8Y1K9 A0A3Q2PSI8 A0A1L8EDA3 A0A3P9JY68 E9J331 B4K0F6 A0A3Q0RLG6 A0A1A8EY25 A0A0M3QUM4 A0A1A7ZLU5 A0A3Q1J9L5 A0A3P8U4V6 A0A1I8MIU5 A0A1A8JQ36 A0A1A8CW35 A0A3B4XE96 A0A3B4V057 A0A1L8ES41 A0A2U9BEW4 A0A3B3QPR3 A0A3Q1CBD9 A0A293N4Q0 A0A3B1JW97 A0A1A8SIU3 A0A1A8MN39 A0A1A7X9K4 V5HK20 A0A3S2N4J7 Q6GR45 H2YS09 B4LIY3 A0A1I8PD35 A0A2I4BSR8 C1BL53 W5MM18
A0A2A4JS04 A0A194PSS3 A0A3S2NZ78 A0A194RD87 S4P457 A0A088A2P5 E0VVM1 A0A0V0G988 A0A2A3EH32 A0A1S4EIA4 A0A067QMJ9 K7IPC1 A0A0P4W221 R4G4M6 A0A224XV59 A0A023F9D2 E2BSI5 R4WJE1 A0A2R7W8G1 A0A023GGS0 A0A158NE26 A0A1B6DEM1 G3MMH3 A0A1B6G0I8 A0A1B6IX34 A0A023FXX0 A0A1E1XIB0 A0A131XI39 A0A224Z9Z6 A0A131Z3M0 L7M4J9 A0A023FGL5 A0A0C9S1Z6 A0A0K8TRQ2 A0A1B6K9R5 F4W519 A0A0A9YU28 A0A2P6LBR9 A0A195BQL5 A0A195F545 A0A151XED4 A0A151IF09 A0A0L7RB36 A0A087U4V8 A0A195D9U9 A0A2R5LF86 A0A131XUQ6 A0A026WZX9 A0A1Z5LHA1 H2LCJ9 A0A3P8WUI6 A0A3B5BCR8 E2AQP0 A0A3B4DSN8 A0A1Y1L5Y5 I5APZ0 A0A3Q3LQP7 A0A3B3BAX9 A0A0Q9XAB8 A0A0P8Y1K9 A0A3Q2PSI8 A0A1L8EDA3 A0A3P9JY68 E9J331 B4K0F6 A0A3Q0RLG6 A0A1A8EY25 A0A0M3QUM4 A0A1A7ZLU5 A0A3Q1J9L5 A0A3P8U4V6 A0A1I8MIU5 A0A1A8JQ36 A0A1A8CW35 A0A3B4XE96 A0A3B4V057 A0A1L8ES41 A0A2U9BEW4 A0A3B3QPR3 A0A3Q1CBD9 A0A293N4Q0 A0A3B1JW97 A0A1A8SIU3 A0A1A8MN39 A0A1A7X9K4 V5HK20 A0A3S2N4J7 Q6GR45 H2YS09 B4LIY3 A0A1I8PD35 A0A2I4BSR8 C1BL53 W5MM18
PDB
5T62
E-value=3.20737e-101,
Score=938
Ontologies
KEGG
GO
PANTHER
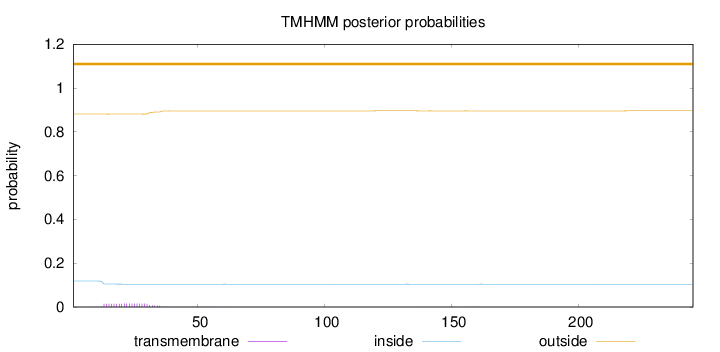
Topology
Subcellular location
Cytoplasm
Shuttles between cytoplasm and nucleus/nucleolus. With evidence from 1 publications.
Nucleus Shuttles between cytoplasm and nucleus/nucleolus. With evidence from 1 publications.
Nucleolus Shuttles between cytoplasm and nucleus/nucleolus. With evidence from 1 publications.
Nucleus Shuttles between cytoplasm and nucleus/nucleolus. With evidence from 1 publications.
Nucleolus Shuttles between cytoplasm and nucleus/nucleolus. With evidence from 1 publications.
Length:
245
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.33643
Exp number, first 60 AAs:
0.321
Total prob of N-in:
0.11876
outside
1 - 245
Population Genetic Test Statistics
Pi
13.278972
Theta
12.867727
Tajima's D
0.130474
CLR
0
CSRT
0.408879556022199
Interpretation
Uncertain
