Pre Gene Modal
BGIBMGA009918
Annotation
PREDICTED:_ras-related_protein_Ral-a_isoform_X2_[Bombyx_mori]
Full name
Ras-related protein Ral-a
Location in the cell
Cytoplasmic Reliability : 1.932 Nuclear Reliability : 1.836
Sequence
CDS
ATGGGTGGACGATCTCACAGCCCGCCTGGTGCTAAGTCAGCCATAACGTACGCATTCATTTTATGTAATGTCCAGGTTGTCCTGGATGGAGAGGAAGTCCAGATCGATATCCTGGACACAGCGGGCCAGGAGGACTATGCCGCCATCAGGGACAACTACTTTAGATCCGGCGAGGGCTTCCTTTGCGTTTTCTCCATCACTGAGCCTGAGAGCTTCGATGCCACACAGGAGTTCAGAGAGCAGATCCTTCGTGTTAAAAACGATGACAACATTCCGTTCCTGCTGGTCGGGAACAAGTCGGACCTCGCGGACAAGCGCCGGGTGCCGCTCGAGACCTGCAGGGAGAGGGCGCACAACTGGCAGGTGCCCTACGTCGAGACCTCGGCGAAAACTAGGGACAATGTCGATAAGGTGTTCTTCGACCTGATGAGGGAGATCCGCTCGCGGAAGTCTGACGACAGCCGCGCGACCAACGGCGAGGGAGGGAAGGGGAAAAAGAGCAAGAAATTCAAATGTGTCATATTGTAG
Protein
MGGRSHSPPGAKSAITYAFILCNVQVVLDGEEVQIDILDTAGQEDYAAIRDNYFRSGEGFLCVFSITEPESFDATQEFREQILRVKNDDNIPFLLVGNKSDLADKRRVPLETCRERAHNWQVPYVETSAKTRDNVDKVFFDLMREIRSRKSDDSRATNGEGGKGKKSKKFKCVIL
Summary
Similarity
Belongs to the small GTPase superfamily. Ras family.
Keywords
3D-structure
Cell membrane
Complete proteome
GTP-binding
Lipoprotein
Membrane
Methylation
Nucleotide-binding
Prenylation
Reference proteome
Feature
chain Ras-related protein Ral-a
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
H9JK67
A0A194RDA3
A0A2H1WCR1
A0A1E1VYX1
A0A194PQR6
S4PFR7
+ More
A0A212FEN5 W8B7H6 A0A034V2V4 A0A0K8V4T4 A0A0A1X0A1 A0A034V4Z5 A0A2M4CT93 A0A2M4AYD1 A0A1Y9G860 A0A2M4C056 A0A2M4C008 W5J225 B3MW90 A0A0P8Y8F7 A0A1W4V381 A0A1Q3F7P6 B0WYZ7 U5EWZ6 A0A0M4F8F7 A0A0K8V5R9 A0A2M4CWC3 A0A1I8NTH8 Q16FH9 A0A023EJD7 A0A182LWQ5 A0A182RAH8 A0A182N3R7 A0A182TW39 A0A182KT84 A0A182HPL6 A0A084WIY7 A0A182IPD4 Q7PKD8 B4NDD6 B4M1L5 B4L4V9 A0A2M4CTL8 B3NSR1 B4Q0I5 P48555 A0A3B0JXT0 Q29GN2 A0A2M4CTB5 A0A182X063 B4JN35 A0A182P1F9 A0A1A9W416 B4R469 B4I156 A0A1I8MZ99 A0A182WD37 A0A1L8DME0 A0A1B0DH23 A0A182JZV2 A0A2M4AZ62 A0A182V2M1 A0A3L8E074 A0A0L7QV18 A0A1B6KRI4 A0A1J1IY57 A0A1B6FUQ4 A0A1B6IHK5 A0A195CN21 A0A195DA23 A0A195F3I9 A0A151XEM3 A0A195BP69 A0A158NDM7 A0A0J7K3H2 A0A2M3ZDF6 E9IYV4 A0A224XX43 A0A0P4VYD8 T1HN74 A0A069DPZ8 A0A182F2S4 A0A2M4C045 A0A2M4AYG4 W5JEW5 A0A023FAN0 A0A0A9XGY7 R4G589 A0A0L7LHQ6 A0A154PTX6 A0A2M3ZK58 A0A2M3ZDJ0 A0A087ZWD6 A0A2R7X285 J3JYW1 A0A2S2R8S7 J9JYL2 A0A2S2NIW1 A0A1B6CQ98
A0A212FEN5 W8B7H6 A0A034V2V4 A0A0K8V4T4 A0A0A1X0A1 A0A034V4Z5 A0A2M4CT93 A0A2M4AYD1 A0A1Y9G860 A0A2M4C056 A0A2M4C008 W5J225 B3MW90 A0A0P8Y8F7 A0A1W4V381 A0A1Q3F7P6 B0WYZ7 U5EWZ6 A0A0M4F8F7 A0A0K8V5R9 A0A2M4CWC3 A0A1I8NTH8 Q16FH9 A0A023EJD7 A0A182LWQ5 A0A182RAH8 A0A182N3R7 A0A182TW39 A0A182KT84 A0A182HPL6 A0A084WIY7 A0A182IPD4 Q7PKD8 B4NDD6 B4M1L5 B4L4V9 A0A2M4CTL8 B3NSR1 B4Q0I5 P48555 A0A3B0JXT0 Q29GN2 A0A2M4CTB5 A0A182X063 B4JN35 A0A182P1F9 A0A1A9W416 B4R469 B4I156 A0A1I8MZ99 A0A182WD37 A0A1L8DME0 A0A1B0DH23 A0A182JZV2 A0A2M4AZ62 A0A182V2M1 A0A3L8E074 A0A0L7QV18 A0A1B6KRI4 A0A1J1IY57 A0A1B6FUQ4 A0A1B6IHK5 A0A195CN21 A0A195DA23 A0A195F3I9 A0A151XEM3 A0A195BP69 A0A158NDM7 A0A0J7K3H2 A0A2M3ZDF6 E9IYV4 A0A224XX43 A0A0P4VYD8 T1HN74 A0A069DPZ8 A0A182F2S4 A0A2M4C045 A0A2M4AYG4 W5JEW5 A0A023FAN0 A0A0A9XGY7 R4G589 A0A0L7LHQ6 A0A154PTX6 A0A2M3ZK58 A0A2M3ZDJ0 A0A087ZWD6 A0A2R7X285 J3JYW1 A0A2S2R8S7 J9JYL2 A0A2S2NIW1 A0A1B6CQ98
Pubmed
19121390
26354079
23622113
22118469
24495485
25348373
+ More
25830018 20920257 23761445 17994087 17510324 24945155 26483478 20966253 24438588 12364791 18057021 17550304 10731132 12537572 12537569 15632085 25315136 30249741 21347285 21282665 27129103 26334808 25474469 25401762 26823975 26227816 22516182 23537049
25830018 20920257 23761445 17994087 17510324 24945155 26483478 20966253 24438588 12364791 18057021 17550304 10731132 12537572 12537569 15632085 25315136 30249741 21347285 21282665 27129103 26334808 25474469 25401762 26823975 26227816 22516182 23537049
EMBL
BABH01026737
KQ460367
KPJ15449.1
ODYU01007465
SOQ50264.1
GDQN01011183
+ More
GDQN01006823 JAT79871.1 JAT84231.1 KQ459595 KPI95791.1 GAIX01006450 JAA86110.1 AGBW02008930 OWR52157.1 GAMC01017364 JAB89191.1 GAKP01022113 JAC36839.1 GDHF01018581 JAI33733.1 GBXI01011829 GBXI01010209 JAD02463.1 JAD04083.1 GAKP01022118 GAKP01022117 GAKP01022116 JAC36835.1 GGFL01004362 MBW68540.1 GGFK01012476 MBW45797.1 GGFJ01009513 MBW58654.1 GGFJ01009514 MBW58655.1 ADMH02002211 ETN57786.1 CH902625 EDV35235.2 KPU75489.1 GFDL01011460 JAV23585.1 DS232201 EDS37243.1 GANO01001182 JAB58689.1 CP012528 ALC48282.1 GDHF01018055 GDHF01016344 JAI34259.1 JAI35970.1 GGFL01004970 MBW69148.1 CH478433 EAT32994.1 JXUM01030924 JXUM01030925 GAPW01004538 KQ560902 JAC09060.1 KXJ80418.1 AXCM01006031 APCN01003265 ATLV01023954 KE525347 KFB50181.1 AXCP01008560 AAAB01008986 EAA43300.3 CH964239 EDW82842.2 CH940651 EDW65569.2 CH933810 EDW07587.2 GGFL01004343 MBW68521.1 CH954180 EDV45741.2 KQS29686.1 CM000162 EDX01269.2 KRK06040.1 U23800 AE014298 AY118912 OUUW01000003 SPP78166.1 CH379064 EAL32077.1 GGFL01004414 MBW68592.1 CH916371 EDV92128.1 CM000366 EDX17011.1 CH480819 EDW53237.1 GFDF01006547 JAV07537.1 AJVK01015314 AJVK01015315 AJVK01015316 GGFK01012760 MBW46081.1 QOIP01000002 RLU26104.1 KQ414727 KOC62483.1 GEBQ01025908 JAT14069.1 CVRI01000064 CRL05024.1 GECZ01016046 JAS53723.1 GECU01036392 GECU01032281 GECU01023839 GECU01021316 GECU01008697 GECU01005893 JAS71314.1 JAS75425.1 JAS83867.1 JAS86390.1 JAS99009.1 JAT01814.1 KQ977600 KYN01484.1 KQ981082 KYN09712.1 KQ981820 KYN35145.1 KQ982254 KYQ58781.1 KQ976424 KYM88312.1 ADTU01012715 ADTU01012716 LBMM01014986 KMQ84973.1 GGFM01005778 MBW26529.1 GL767051 EFZ14247.1 GFTR01003356 JAW13070.1 GDKW01000956 JAI55639.1 ACPB03007651 GBGD01002999 JAC85890.1 GGFJ01009511 MBW58652.1 GGFK01012499 MBW45820.1 ADMH02001599 ETN61888.1 GBBI01000623 JAC18089.1 GBHO01023617 GBRD01017354 GDHC01012832 GDHC01007932 JAG19987.1 JAG48473.1 JAQ05797.1 JAQ10697.1 GAHY01000725 JAA76785.1 JTDY01001026 KOB75093.1 KQ435127 KZC14808.1 GGFM01008170 MBW28921.1 GGFM01005828 MBW26579.1 KK856496 PTY25903.1 APGK01029531 APGK01029532 BT128442 KB740735 KB631642 AEE63399.1 ENN79183.1 ERL84910.1 GGMS01017151 MBY86354.1 ABLF02040827 GGMR01004480 MBY17099.1 GEDC01021737 JAS15561.1
GDQN01006823 JAT79871.1 JAT84231.1 KQ459595 KPI95791.1 GAIX01006450 JAA86110.1 AGBW02008930 OWR52157.1 GAMC01017364 JAB89191.1 GAKP01022113 JAC36839.1 GDHF01018581 JAI33733.1 GBXI01011829 GBXI01010209 JAD02463.1 JAD04083.1 GAKP01022118 GAKP01022117 GAKP01022116 JAC36835.1 GGFL01004362 MBW68540.1 GGFK01012476 MBW45797.1 GGFJ01009513 MBW58654.1 GGFJ01009514 MBW58655.1 ADMH02002211 ETN57786.1 CH902625 EDV35235.2 KPU75489.1 GFDL01011460 JAV23585.1 DS232201 EDS37243.1 GANO01001182 JAB58689.1 CP012528 ALC48282.1 GDHF01018055 GDHF01016344 JAI34259.1 JAI35970.1 GGFL01004970 MBW69148.1 CH478433 EAT32994.1 JXUM01030924 JXUM01030925 GAPW01004538 KQ560902 JAC09060.1 KXJ80418.1 AXCM01006031 APCN01003265 ATLV01023954 KE525347 KFB50181.1 AXCP01008560 AAAB01008986 EAA43300.3 CH964239 EDW82842.2 CH940651 EDW65569.2 CH933810 EDW07587.2 GGFL01004343 MBW68521.1 CH954180 EDV45741.2 KQS29686.1 CM000162 EDX01269.2 KRK06040.1 U23800 AE014298 AY118912 OUUW01000003 SPP78166.1 CH379064 EAL32077.1 GGFL01004414 MBW68592.1 CH916371 EDV92128.1 CM000366 EDX17011.1 CH480819 EDW53237.1 GFDF01006547 JAV07537.1 AJVK01015314 AJVK01015315 AJVK01015316 GGFK01012760 MBW46081.1 QOIP01000002 RLU26104.1 KQ414727 KOC62483.1 GEBQ01025908 JAT14069.1 CVRI01000064 CRL05024.1 GECZ01016046 JAS53723.1 GECU01036392 GECU01032281 GECU01023839 GECU01021316 GECU01008697 GECU01005893 JAS71314.1 JAS75425.1 JAS83867.1 JAS86390.1 JAS99009.1 JAT01814.1 KQ977600 KYN01484.1 KQ981082 KYN09712.1 KQ981820 KYN35145.1 KQ982254 KYQ58781.1 KQ976424 KYM88312.1 ADTU01012715 ADTU01012716 LBMM01014986 KMQ84973.1 GGFM01005778 MBW26529.1 GL767051 EFZ14247.1 GFTR01003356 JAW13070.1 GDKW01000956 JAI55639.1 ACPB03007651 GBGD01002999 JAC85890.1 GGFJ01009511 MBW58652.1 GGFK01012499 MBW45820.1 ADMH02001599 ETN61888.1 GBBI01000623 JAC18089.1 GBHO01023617 GBRD01017354 GDHC01012832 GDHC01007932 JAG19987.1 JAG48473.1 JAQ05797.1 JAQ10697.1 GAHY01000725 JAA76785.1 JTDY01001026 KOB75093.1 KQ435127 KZC14808.1 GGFM01008170 MBW28921.1 GGFM01005828 MBW26579.1 KK856496 PTY25903.1 APGK01029531 APGK01029532 BT128442 KB740735 KB631642 AEE63399.1 ENN79183.1 ERL84910.1 GGMS01017151 MBY86354.1 ABLF02040827 GGMR01004480 MBY17099.1 GEDC01021737 JAS15561.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000069272
UP000000673
+ More
UP000007801 UP000192221 UP000002320 UP000092553 UP000095300 UP000008820 UP000069940 UP000249989 UP000075883 UP000075900 UP000075884 UP000075902 UP000075882 UP000075840 UP000030765 UP000075880 UP000007062 UP000007798 UP000008792 UP000009192 UP000008711 UP000002282 UP000000803 UP000268350 UP000001819 UP000076407 UP000001070 UP000075885 UP000091820 UP000000304 UP000001292 UP000095301 UP000075920 UP000092462 UP000075881 UP000075903 UP000279307 UP000053825 UP000183832 UP000078542 UP000078492 UP000078541 UP000075809 UP000078540 UP000005205 UP000036403 UP000015103 UP000037510 UP000076502 UP000005203 UP000019118 UP000030742 UP000007819
UP000007801 UP000192221 UP000002320 UP000092553 UP000095300 UP000008820 UP000069940 UP000249989 UP000075883 UP000075900 UP000075884 UP000075902 UP000075882 UP000075840 UP000030765 UP000075880 UP000007062 UP000007798 UP000008792 UP000009192 UP000008711 UP000002282 UP000000803 UP000268350 UP000001819 UP000076407 UP000001070 UP000075885 UP000091820 UP000000304 UP000001292 UP000095301 UP000075920 UP000092462 UP000075881 UP000075903 UP000279307 UP000053825 UP000183832 UP000078542 UP000078492 UP000078541 UP000075809 UP000078540 UP000005205 UP000036403 UP000015103 UP000037510 UP000076502 UP000005203 UP000019118 UP000030742 UP000007819
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JK67
A0A194RDA3
A0A2H1WCR1
A0A1E1VYX1
A0A194PQR6
S4PFR7
+ More
A0A212FEN5 W8B7H6 A0A034V2V4 A0A0K8V4T4 A0A0A1X0A1 A0A034V4Z5 A0A2M4CT93 A0A2M4AYD1 A0A1Y9G860 A0A2M4C056 A0A2M4C008 W5J225 B3MW90 A0A0P8Y8F7 A0A1W4V381 A0A1Q3F7P6 B0WYZ7 U5EWZ6 A0A0M4F8F7 A0A0K8V5R9 A0A2M4CWC3 A0A1I8NTH8 Q16FH9 A0A023EJD7 A0A182LWQ5 A0A182RAH8 A0A182N3R7 A0A182TW39 A0A182KT84 A0A182HPL6 A0A084WIY7 A0A182IPD4 Q7PKD8 B4NDD6 B4M1L5 B4L4V9 A0A2M4CTL8 B3NSR1 B4Q0I5 P48555 A0A3B0JXT0 Q29GN2 A0A2M4CTB5 A0A182X063 B4JN35 A0A182P1F9 A0A1A9W416 B4R469 B4I156 A0A1I8MZ99 A0A182WD37 A0A1L8DME0 A0A1B0DH23 A0A182JZV2 A0A2M4AZ62 A0A182V2M1 A0A3L8E074 A0A0L7QV18 A0A1B6KRI4 A0A1J1IY57 A0A1B6FUQ4 A0A1B6IHK5 A0A195CN21 A0A195DA23 A0A195F3I9 A0A151XEM3 A0A195BP69 A0A158NDM7 A0A0J7K3H2 A0A2M3ZDF6 E9IYV4 A0A224XX43 A0A0P4VYD8 T1HN74 A0A069DPZ8 A0A182F2S4 A0A2M4C045 A0A2M4AYG4 W5JEW5 A0A023FAN0 A0A0A9XGY7 R4G589 A0A0L7LHQ6 A0A154PTX6 A0A2M3ZK58 A0A2M3ZDJ0 A0A087ZWD6 A0A2R7X285 J3JYW1 A0A2S2R8S7 J9JYL2 A0A2S2NIW1 A0A1B6CQ98
A0A212FEN5 W8B7H6 A0A034V2V4 A0A0K8V4T4 A0A0A1X0A1 A0A034V4Z5 A0A2M4CT93 A0A2M4AYD1 A0A1Y9G860 A0A2M4C056 A0A2M4C008 W5J225 B3MW90 A0A0P8Y8F7 A0A1W4V381 A0A1Q3F7P6 B0WYZ7 U5EWZ6 A0A0M4F8F7 A0A0K8V5R9 A0A2M4CWC3 A0A1I8NTH8 Q16FH9 A0A023EJD7 A0A182LWQ5 A0A182RAH8 A0A182N3R7 A0A182TW39 A0A182KT84 A0A182HPL6 A0A084WIY7 A0A182IPD4 Q7PKD8 B4NDD6 B4M1L5 B4L4V9 A0A2M4CTL8 B3NSR1 B4Q0I5 P48555 A0A3B0JXT0 Q29GN2 A0A2M4CTB5 A0A182X063 B4JN35 A0A182P1F9 A0A1A9W416 B4R469 B4I156 A0A1I8MZ99 A0A182WD37 A0A1L8DME0 A0A1B0DH23 A0A182JZV2 A0A2M4AZ62 A0A182V2M1 A0A3L8E074 A0A0L7QV18 A0A1B6KRI4 A0A1J1IY57 A0A1B6FUQ4 A0A1B6IHK5 A0A195CN21 A0A195DA23 A0A195F3I9 A0A151XEM3 A0A195BP69 A0A158NDM7 A0A0J7K3H2 A0A2M3ZDF6 E9IYV4 A0A224XX43 A0A0P4VYD8 T1HN74 A0A069DPZ8 A0A182F2S4 A0A2M4C045 A0A2M4AYG4 W5JEW5 A0A023FAN0 A0A0A9XGY7 R4G589 A0A0L7LHQ6 A0A154PTX6 A0A2M3ZK58 A0A2M3ZDJ0 A0A087ZWD6 A0A2R7X285 J3JYW1 A0A2S2R8S7 J9JYL2 A0A2S2NIW1 A0A1B6CQ98
PDB
5CM9
E-value=1.22095e-60,
Score=586
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
Cleavage furrow
Midbody
Midbody ring
Cleavage furrow
Midbody
Midbody ring
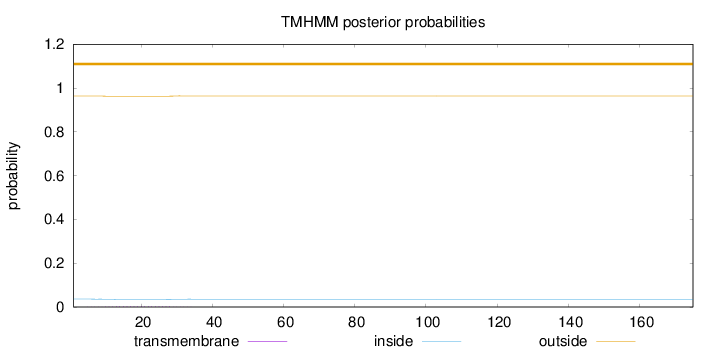
Length:
175
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06103
Exp number, first 60 AAs:
0.06103
Total prob of N-in:
0.03618
outside
1 - 175
Population Genetic Test Statistics
Pi
36.47348
Theta
25.823992
Tajima's D
1.369927
CLR
0.656865
CSRT
0.764061796910155
Interpretation
Uncertain
