Gene
KWMTBOMO04817
Pre Gene Modal
BGIBMGA009832
Annotation
PREDICTED:_WD_repeat-containing_protein_mio_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.515 Nuclear Reliability : 1.002
Sequence
CDS
ATGTCTAGCAGATTGGATATATTATGGTCACCGGCACATCATGACAAATTCATTGTTTGGGGCCCAGACATAACCTTGTACGAAGTGGCAAGGATAAAAGAAATCGAGAAGAAAACGACTTTCACTCCAATTTCCTCAACCTATGGAGCCACAGTTGTTGCGTCCCAGAGTGCTAGTGGAGTGCGGTGCGTTGATATCAGTGCTGAACCAGAGCAGCCTGAGCCATTGCTCGCTCTTGGATATGCTAGTGGACGGGTCTCACTTACCACCCTTAAACAAACATATGACCCTTTGGGACTAGTTGGAAGAGAGTTTGCACCAAGATACCAAAGGCTTTGCAACTCTGTGTCATGGAGTCACATGGAGACAAACATGTTGGCAGTCGCGATGGATAAACATCGTAGTGACCACTGCATACTTGTATGGGATGTAAACCGAGGTTCGGCAGCTGCTGATGAGGCTTTAGAAAGTACTTCTCAAGGAAGTGTAGATCCGATGCGACCACTAGCAGAAATGGGACTATCTGAGACAGCACATAATGTAACCTGGTCTAACTGCTCGAACAGGACCTTATTGGCTTCAATGAACTTGAAGTACATCAAAATATTTGATCTTAGAGATTTATCGAACAAAGCCGTGAGCATGACCGCGACCCGCTACTGCTACGGTGTGTGTGCCGACCCGAACAGCTCCCACCAGCTCGCCTCCCGCGGAGAGAACGTGGTCTGTCTTTGGGACACCAGGCACTTCGACCGCCCCGTACTCTCTCTGCAGCATCCCCGGCCTCTAGCCGTCACCCATCTGCAGTGGTGCCCTACCAGACGCAACCTTCTTCTCTCGCTACAGAGAGATTCCAAGTCGCTCCGCATCCACGACATACAGCAGGCCAGTGCGGAGTGGAAGAAAAACGCGCTCGATGTGCTGACGGCGACGGCCGTAGAGGCCGAAGTGGAGGAGTACTCTCGCAGACCGAGCGTGTTGGAGCGTGACGTCACTCCTGGAGGGTCGCAGCCGCTCGTGGCCTTCTGCTGCCATCCGGCTCACGAGGCGCGTCTGCTGGCACTCAGCGCCACAGGCGGTCTGGTGGACTACACGGTCTGCGAGCGCGTGGCGGTGTCCTGGGGGGCGGGGGGAGGGCTGGCCTGGTCCGGGGGAGGAGGGGGACTCCGTGTCCTCAACGACGACTTGTGCGGGGTCGTGAAGGACATCTCGCACGTCATGAGACGACGCGCCCAAACCGACTACGGCCTCAAGCAAGACCTGTGGCAGAACGCAGACCTCGCCGATGACGACGCGCTCAGTGCCCTGTGGCACTTCATGGCCTTGAGCAAGTCCTTGGTGGAAGATGGTTGCATAAGGAACAGTCCGTGGAAGCACCCGGGCGTGCTGACCGTGCTGCGGTCCCCGGGCGACGGCGGCTACCGCTCCGAGATAGTGCCCTCCCTGCTGCCCGATATGCCTAACCGCAGGGTCACGCTCTACAGGAGTGCAGAGCGAACTCGTGCACTGCAGCTGTGCGGGTGGGGGTGGGGGTGGGAGAACGCGGACGCGGGGGTGGAGCGCGCCGAGGCCGAGGGCACCCCGTGTCGCGCCGCCGCACTCGCCGCCTTCCACCTCCGGCTTAGGACTGCTCTGGACGTGCTGGCCCGGGCGCACGAGCCGCAGAATTGTAACCTGTGCAAGATCCAATTCAGTGATGGTGTTCAATGCGGTACGTGCAAGAAGCATTTTGACTTCGGATGCGCTAATATTACAGAAGCTGGATATAGAAAACTGGGCCCCGAAAGAAGGGCGGCATGGAAGTGTTCTGCTTGCAGAAACCCTTCGCAAGCGGTTGCTTCACAGGCCAGCCCAGTCTCTCCAGTTCAGGGCTTAGTGACGCTAGAAATGATCCACGCTGAATTATGCGGCGTAAATTGA
Protein
MSSRLDILWSPAHHDKFIVWGPDITLYEVARIKEIEKKTTFTPISSTYGATVVASQSASGVRCVDISAEPEQPEPLLALGYASGRVSLTTLKQTYDPLGLVGREFAPRYQRLCNSVSWSHMETNMLAVAMDKHRSDHCILVWDVNRGSAAADEALESTSQGSVDPMRPLAEMGLSETAHNVTWSNCSNRTLLASMNLKYIKIFDLRDLSNKAVSMTATRYCYGVCADPNSSHQLASRGENVVCLWDTRHFDRPVLSLQHPRPLAVTHLQWCPTRRNLLLSLQRDSKSLRIHDIQQASAEWKKNALDVLTATAVEAEVEEYSRRPSVLERDVTPGGSQPLVAFCCHPAHEARLLALSATGGLVDYTVCERVAVSWGAGGGLAWSGGGGGLRVLNDDLCGVVKDISHVMRRRAQTDYGLKQDLWQNADLADDDALSALWHFMALSKSLVEDGCIRNSPWKHPGVLTVLRSPGDGGYRSEIVPSLLPDMPNRRVTLYRSAERTRALQLCGWGWGWENADAGVERAEAEGTPCRAAALAAFHLRLRTALDVLARAHEPQNCNLCKIQFSDGVQCGTCKKHFDFGCANITEAGYRKLGPERRAAWKCSACRNPSQAVASQASPVSPVQGLVTLEMIHAELCGVN
Summary
Uniprot
A0A2H1WB06
A0A2A4K6R2
A0A2A4K7L7
A0A194PR54
A0A194RC80
A0A212FEH8
+ More
A0A067R031 A0A2J7R195 A0A2J7R1C2 A0A2J7R1A2 A0A1B6E123 A0A1B6CGC5 A0A1B6L8L7 A0A1B6IDD1 A0A1B6H0J4 A0A0C9QYF2 A0A310SSH7 A0A154PSS2 A0A087ZWD8 A0A0L7QV95 A0A1Y1KB36 A0A0N0BEV0 A0A2A3ELD9 D6X052 A0A1W4XSV9 E2BJ86 A0A232FKD7 A0A0P4VU12 A0A139WCL5 A0A023F548 K7IYZ6 A0A026W3K6 A0A3L8DLU9 A0A0A9ZBQ5 A0A0A9ZA57 A0A146LXW2 T1HA87 A0A0J7NZZ8 A0A158NDY1 A0A195BPP5 F4X1U9 A0A151X4E6 A0A195D806 A0A195ERF4 E9IGT8 A0A0T6B5S4 E0VU36 A0A2S2RA21 A0A0N7ZBK5 A0A195C6Y1 C3YUN5 A0A3R7PU50 E2A9M0 C3Z8N1 A0A1S3ILI1 A0A1S3ILZ4 X1WJ75 A0A1S3INI8 A0A1Z5KWS2 A0A1D2NKF9 A0A131Y0V4 A0A0P5V8E2 A0A210PU67 A0A2R5LMG4 A0A090X8F8 A0A3P9I9E0 A0A293LWX7 A0A0P5FW72 A0A0N8CLM2 A0A0P5D9N6 A0A0P5ZGA0 A0A0P6BDA7 A0A3P9KR43 H2MPQ3 A0A3P9CET6 A0A0N8C529 I3JH67 A0A0N8CAH9 A0A2T7PAB9 W5MZ29 A0A3B4H0B2 A0A3Q2X1F3 A0A3P8P0G1 A0A0P6FCQ2 A0A0P5RM25 A0A3Q1GPE2 A0A0P6DJD4 A0A0P6DTT7 A0A0P6FKK7 A0A0P5PX41 A0A3Q1HKV4 A0A0P6FJ47 A0A0P5MDJ9
A0A067R031 A0A2J7R195 A0A2J7R1C2 A0A2J7R1A2 A0A1B6E123 A0A1B6CGC5 A0A1B6L8L7 A0A1B6IDD1 A0A1B6H0J4 A0A0C9QYF2 A0A310SSH7 A0A154PSS2 A0A087ZWD8 A0A0L7QV95 A0A1Y1KB36 A0A0N0BEV0 A0A2A3ELD9 D6X052 A0A1W4XSV9 E2BJ86 A0A232FKD7 A0A0P4VU12 A0A139WCL5 A0A023F548 K7IYZ6 A0A026W3K6 A0A3L8DLU9 A0A0A9ZBQ5 A0A0A9ZA57 A0A146LXW2 T1HA87 A0A0J7NZZ8 A0A158NDY1 A0A195BPP5 F4X1U9 A0A151X4E6 A0A195D806 A0A195ERF4 E9IGT8 A0A0T6B5S4 E0VU36 A0A2S2RA21 A0A0N7ZBK5 A0A195C6Y1 C3YUN5 A0A3R7PU50 E2A9M0 C3Z8N1 A0A1S3ILI1 A0A1S3ILZ4 X1WJ75 A0A1S3INI8 A0A1Z5KWS2 A0A1D2NKF9 A0A131Y0V4 A0A0P5V8E2 A0A210PU67 A0A2R5LMG4 A0A090X8F8 A0A3P9I9E0 A0A293LWX7 A0A0P5FW72 A0A0N8CLM2 A0A0P5D9N6 A0A0P5ZGA0 A0A0P6BDA7 A0A3P9KR43 H2MPQ3 A0A3P9CET6 A0A0N8C529 I3JH67 A0A0N8CAH9 A0A2T7PAB9 W5MZ29 A0A3B4H0B2 A0A3Q2X1F3 A0A3P8P0G1 A0A0P6FCQ2 A0A0P5RM25 A0A3Q1GPE2 A0A0P6DJD4 A0A0P6DTT7 A0A0P6FKK7 A0A0P5PX41 A0A3Q1HKV4 A0A0P6FJ47 A0A0P5MDJ9
Pubmed
EMBL
ODYU01007465
SOQ50265.1
NWSH01000074
PCG79927.1
PCG79928.1
KQ459595
+ More
KPI95792.1 KQ460367 KPJ15448.1 AGBW02008930 OWR52156.1 KK852804 KDR16180.1 NEVH01008208 PNF34611.1 PNF34614.1 PNF34613.1 GEDC01005678 JAS31620.1 GEDC01024751 JAS12547.1 GEBQ01019925 JAT20052.1 GECU01022784 GECU01008675 JAS84922.1 JAS99031.1 GECZ01001584 JAS68185.1 GBYB01000729 GBYB01000730 JAG70496.1 JAG70497.1 KQ759851 OAD62560.1 KQ435127 KZC14807.1 KQ414727 KOC62484.1 GEZM01094054 GEZM01094053 JAV55967.1 KQ435821 KOX72418.1 KZ288215 PBC32538.1 KQ971371 EFA10726.1 GL448558 EFN84236.1 NNAY01000120 OXU30787.1 GDKW01001086 JAI55509.1 KQ971365 KYB25657.1 GBBI01002041 JAC16671.1 KK107503 EZA49619.1 QOIP01000006 RLU21193.1 GBHO01002273 JAG41331.1 GBHO01002275 JAG41329.1 GDHC01006226 JAQ12403.1 ACPB03021149 LBMM01000604 KMQ97975.1 ADTU01013072 KQ976424 KYM88492.1 GL888551 EGI59606.1 KQ982548 KYQ55293.1 KQ981153 KYN08987.1 KQ981993 KYN30803.1 GL763054 EFZ20281.1 LJIG01009610 KRT82735.1 DS235779 EEB16892.1 GGMS01017632 MBY86835.1 GDRN01082118 GDRN01082117 JAI61904.1 KQ978251 KYM95923.1 GG666554 EEN55936.1 QCYY01001566 ROT77125.1 GL437918 EFN69822.1 GG666594 EEN51136.1 ABLF02029453 GFJQ02007413 JAV99556.1 LJIJ01000022 ODN05456.1 GEFM01004285 JAP71511.1 GDIP01105628 GDIP01105627 JAL98086.1 NEDP02005493 OWF40005.1 GGLE01006568 MBY10694.1 GBIH01002741 JAC91969.1 GFWV01013248 MAA37977.1 GDIQ01253134 JAJ98590.1 GDIP01119671 JAL84043.1 GDIP01164544 JAJ58858.1 GDIP01045044 LRGB01000389 JAM58671.1 KZS19408.1 GDIP01030948 JAM72767.1 GDIQ01114027 JAL37699.1 AERX01023930 GDIQ01098827 JAL52899.1 PZQS01000005 PVD30358.1 AHAT01004624 GDIQ01061891 JAN32846.1 GDIQ01098826 JAL52900.1 GDIQ01076633 JAN18104.1 GDIQ01076634 JAN18103.1 GDIQ01048157 JAN46580.1 GDIQ01122176 JAL29550.1 GDIQ01063600 JAN31137.1 GDIQ01165415 JAK86310.1
KPI95792.1 KQ460367 KPJ15448.1 AGBW02008930 OWR52156.1 KK852804 KDR16180.1 NEVH01008208 PNF34611.1 PNF34614.1 PNF34613.1 GEDC01005678 JAS31620.1 GEDC01024751 JAS12547.1 GEBQ01019925 JAT20052.1 GECU01022784 GECU01008675 JAS84922.1 JAS99031.1 GECZ01001584 JAS68185.1 GBYB01000729 GBYB01000730 JAG70496.1 JAG70497.1 KQ759851 OAD62560.1 KQ435127 KZC14807.1 KQ414727 KOC62484.1 GEZM01094054 GEZM01094053 JAV55967.1 KQ435821 KOX72418.1 KZ288215 PBC32538.1 KQ971371 EFA10726.1 GL448558 EFN84236.1 NNAY01000120 OXU30787.1 GDKW01001086 JAI55509.1 KQ971365 KYB25657.1 GBBI01002041 JAC16671.1 KK107503 EZA49619.1 QOIP01000006 RLU21193.1 GBHO01002273 JAG41331.1 GBHO01002275 JAG41329.1 GDHC01006226 JAQ12403.1 ACPB03021149 LBMM01000604 KMQ97975.1 ADTU01013072 KQ976424 KYM88492.1 GL888551 EGI59606.1 KQ982548 KYQ55293.1 KQ981153 KYN08987.1 KQ981993 KYN30803.1 GL763054 EFZ20281.1 LJIG01009610 KRT82735.1 DS235779 EEB16892.1 GGMS01017632 MBY86835.1 GDRN01082118 GDRN01082117 JAI61904.1 KQ978251 KYM95923.1 GG666554 EEN55936.1 QCYY01001566 ROT77125.1 GL437918 EFN69822.1 GG666594 EEN51136.1 ABLF02029453 GFJQ02007413 JAV99556.1 LJIJ01000022 ODN05456.1 GEFM01004285 JAP71511.1 GDIP01105628 GDIP01105627 JAL98086.1 NEDP02005493 OWF40005.1 GGLE01006568 MBY10694.1 GBIH01002741 JAC91969.1 GFWV01013248 MAA37977.1 GDIQ01253134 JAJ98590.1 GDIP01119671 JAL84043.1 GDIP01164544 JAJ58858.1 GDIP01045044 LRGB01000389 JAM58671.1 KZS19408.1 GDIP01030948 JAM72767.1 GDIQ01114027 JAL37699.1 AERX01023930 GDIQ01098827 JAL52899.1 PZQS01000005 PVD30358.1 AHAT01004624 GDIQ01061891 JAN32846.1 GDIQ01098826 JAL52900.1 GDIQ01076633 JAN18104.1 GDIQ01076634 JAN18103.1 GDIQ01048157 JAN46580.1 GDIQ01122176 JAL29550.1 GDIQ01063600 JAN31137.1 GDIQ01165415 JAK86310.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000027135
UP000235965
+ More
UP000076502 UP000005203 UP000053825 UP000053105 UP000242457 UP000007266 UP000192223 UP000008237 UP000215335 UP000002358 UP000053097 UP000279307 UP000015103 UP000036403 UP000005205 UP000078540 UP000007755 UP000075809 UP000078492 UP000078541 UP000009046 UP000078542 UP000001554 UP000283509 UP000000311 UP000085678 UP000007819 UP000094527 UP000242188 UP000265200 UP000076858 UP000265180 UP000001038 UP000265160 UP000005207 UP000245119 UP000018468 UP000261460 UP000264840 UP000265100 UP000257200 UP000265040
UP000076502 UP000005203 UP000053825 UP000053105 UP000242457 UP000007266 UP000192223 UP000008237 UP000215335 UP000002358 UP000053097 UP000279307 UP000015103 UP000036403 UP000005205 UP000078540 UP000007755 UP000075809 UP000078492 UP000078541 UP000009046 UP000078542 UP000001554 UP000283509 UP000000311 UP000085678 UP000007819 UP000094527 UP000242188 UP000265200 UP000076858 UP000265180 UP000001038 UP000265160 UP000005207 UP000245119 UP000018468 UP000261460 UP000264840 UP000265100 UP000257200 UP000265040
Interpro
Gene 3D
ProteinModelPortal
A0A2H1WB06
A0A2A4K6R2
A0A2A4K7L7
A0A194PR54
A0A194RC80
A0A212FEH8
+ More
A0A067R031 A0A2J7R195 A0A2J7R1C2 A0A2J7R1A2 A0A1B6E123 A0A1B6CGC5 A0A1B6L8L7 A0A1B6IDD1 A0A1B6H0J4 A0A0C9QYF2 A0A310SSH7 A0A154PSS2 A0A087ZWD8 A0A0L7QV95 A0A1Y1KB36 A0A0N0BEV0 A0A2A3ELD9 D6X052 A0A1W4XSV9 E2BJ86 A0A232FKD7 A0A0P4VU12 A0A139WCL5 A0A023F548 K7IYZ6 A0A026W3K6 A0A3L8DLU9 A0A0A9ZBQ5 A0A0A9ZA57 A0A146LXW2 T1HA87 A0A0J7NZZ8 A0A158NDY1 A0A195BPP5 F4X1U9 A0A151X4E6 A0A195D806 A0A195ERF4 E9IGT8 A0A0T6B5S4 E0VU36 A0A2S2RA21 A0A0N7ZBK5 A0A195C6Y1 C3YUN5 A0A3R7PU50 E2A9M0 C3Z8N1 A0A1S3ILI1 A0A1S3ILZ4 X1WJ75 A0A1S3INI8 A0A1Z5KWS2 A0A1D2NKF9 A0A131Y0V4 A0A0P5V8E2 A0A210PU67 A0A2R5LMG4 A0A090X8F8 A0A3P9I9E0 A0A293LWX7 A0A0P5FW72 A0A0N8CLM2 A0A0P5D9N6 A0A0P5ZGA0 A0A0P6BDA7 A0A3P9KR43 H2MPQ3 A0A3P9CET6 A0A0N8C529 I3JH67 A0A0N8CAH9 A0A2T7PAB9 W5MZ29 A0A3B4H0B2 A0A3Q2X1F3 A0A3P8P0G1 A0A0P6FCQ2 A0A0P5RM25 A0A3Q1GPE2 A0A0P6DJD4 A0A0P6DTT7 A0A0P6FKK7 A0A0P5PX41 A0A3Q1HKV4 A0A0P6FJ47 A0A0P5MDJ9
A0A067R031 A0A2J7R195 A0A2J7R1C2 A0A2J7R1A2 A0A1B6E123 A0A1B6CGC5 A0A1B6L8L7 A0A1B6IDD1 A0A1B6H0J4 A0A0C9QYF2 A0A310SSH7 A0A154PSS2 A0A087ZWD8 A0A0L7QV95 A0A1Y1KB36 A0A0N0BEV0 A0A2A3ELD9 D6X052 A0A1W4XSV9 E2BJ86 A0A232FKD7 A0A0P4VU12 A0A139WCL5 A0A023F548 K7IYZ6 A0A026W3K6 A0A3L8DLU9 A0A0A9ZBQ5 A0A0A9ZA57 A0A146LXW2 T1HA87 A0A0J7NZZ8 A0A158NDY1 A0A195BPP5 F4X1U9 A0A151X4E6 A0A195D806 A0A195ERF4 E9IGT8 A0A0T6B5S4 E0VU36 A0A2S2RA21 A0A0N7ZBK5 A0A195C6Y1 C3YUN5 A0A3R7PU50 E2A9M0 C3Z8N1 A0A1S3ILI1 A0A1S3ILZ4 X1WJ75 A0A1S3INI8 A0A1Z5KWS2 A0A1D2NKF9 A0A131Y0V4 A0A0P5V8E2 A0A210PU67 A0A2R5LMG4 A0A090X8F8 A0A3P9I9E0 A0A293LWX7 A0A0P5FW72 A0A0N8CLM2 A0A0P5D9N6 A0A0P5ZGA0 A0A0P6BDA7 A0A3P9KR43 H2MPQ3 A0A3P9CET6 A0A0N8C529 I3JH67 A0A0N8CAH9 A0A2T7PAB9 W5MZ29 A0A3B4H0B2 A0A3Q2X1F3 A0A3P8P0G1 A0A0P6FCQ2 A0A0P5RM25 A0A3Q1GPE2 A0A0P6DJD4 A0A0P6DTT7 A0A0P6FKK7 A0A0P5PX41 A0A3Q1HKV4 A0A0P6FJ47 A0A0P5MDJ9
PDB
4BZK
E-value=0.00966511,
Score=94
Ontologies
GO
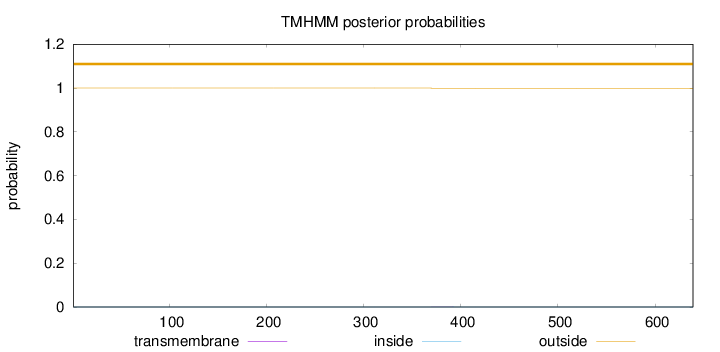
Topology
Length:
639
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00784999999999999
Exp number, first 60 AAs:
0.00168
Total prob of N-in:
0.00030
outside
1 - 639
Population Genetic Test Statistics
Pi
2.929314
Theta
5.123212
Tajima's D
-1.899671
CLR
34.182397
CSRT
0.0173491325433728
Interpretation
Uncertain
