Gene
KWMTBOMO04815
Pre Gene Modal
BGIBMGA009917
Annotation
putative_histone_deacetylase_7A_[Danaus_plexippus]
Full name
Histone deacetylase
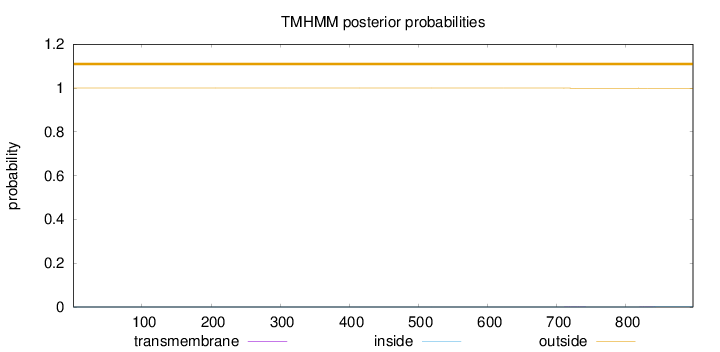
Location in the cell
Nuclear Reliability : 2.599
Sequence
CDS
ATGGCCCACGAGGGCCCAGCAAGGACGGAGGGCTCGCCCCACCACACGCCCTCGCCGCCCCACCTCCCTCACCTCCCAGTCACCGACTCCGGCTTCCATCATCAGATAATGCAGAATCACAGTCCAAGAAAGAATCATATCGTGGAGAGAGCCAAACAAACGTTAGAAAACTCTTTCCTGCAGCAATTGAAGAAGCAGCAACAGCTCCAACAGGAGATTCTACTGCAGCATTTCCAGCAGCAGAGGCAACAGTTGGCAAGAGAACACGAGAACCAGATGAGGGAACATTTGAAGTTATGGGAGCAACAGAAGAAAGCAATGGAGGAGGCAGCGTTGCGCGAGGCCCGAGAGGCGCGGGAGGCTCGGGAGGCCCGGGAGAGGCACGAGCGCGACCGCGTCGAGCTGATGCGGAAGAAGGATATACACGAACACAGCGCCAACGCGTCTACCCAAGTTAAACAGAAATTACAGGAGTTTATAAAAAAGAAACAGGCCAACGCCAATGGCACCGTGACGACTGCCTCCTACCGCAACTGGGGCATAGTGAAGTCTTCATCAGGGGAGTCGATAACGTCGACGGGGGCGACAGCGACGCACCCTTACCGGCTCACGGCGCCGCTGCCACTCAACCTCAACGCGCCCCCTCCGCAACCCTCCCCCGCAGACTACCCCCTGCGCAAGACCGCGTCGGAGCCCAACATGCTGAAGCAGCGGCTGAAAGCGCGCGTCATCGAACGCCGGGCCTCGCCCCTCGCGCGCCGCCCCCTCCGGACCCGGCACAAACACCGCAACTGCGAAGGTGGGTCCCCCCGCGGCTCGCCCCCCGGTGGCTGCGCCGCGGCTCCCATCCGCGAGGAGGAGGAGAACGGCTCCCGCGCCGCAGAGTTCTTGTTTTCGTCTCCGTCACTGCCCAACATCTCTCTGGGACGACCTCACGTGGGTCCCCGCCTGCACCCGCCCCCCGCGGCGCACACCCCGGCGCCCGTGGTGCTGCCCCCCGTGTCGGAGGCGGAGGCGCTGGTGGTGGGCGGGGCGGGGGGCCGGCCGGTGCAGCGCCTCGGTCGCACGCACTCCGCGCCGCTGCCCCTGGGCGACCCGGCCCTGCAGCCGCCCCCCGTGCACCACTACCTCAGAGATCAGATCCGGAAAACTGTGTTGACCCGCGCACATGACGCGGCGGTGCAGCTGCGGGAGGAGGAGGGCGAGGTCATCGACCTGACGTCGAAGCGTAACGATCCTCCCGCCCCCGTCGAGCCGGCCCCCCTCACCCGCGCCCTCTCGTCGCCCCTGGTGGGGGCCAGGGTGCCCGCCACGGGACTCGCCTACGACCCGCTCATGCTCAAGCATGGGTGTGCGTGCGGACAGCACGCCCCCACGCACCCGGAGCACGGCGGCCGCCTGCAGTCCGTGTGGGCGCGCCTGTGCGAGACCGGCCTGGCCGCCCGCACCGAGCGCACGCGGCCCAGGAAGGCCACGCTCGACGAACTGCAGAGCGTGCACACGGAGGCGCACGTGCAGGTGTTCGCGGGGCGGGGGCGCGACGGCGCGGGCGTGGGGGCGGGGGGCGGCGCGGGGGGCGCGGTGCGGCAGCTGGTGCGGCTGGCGTGCGGCGGGCTGGGCGTGGACTCGGACACGGCGTGGTCGGACCTGCACACGGCGCCCGCCGCGCGCCTCGCCGCCGGAGCCCTGCTGGACCTGGCGCTGCGCACCGCGCGCGGACACCTCCGGAACGGGTTCGCGGTGGTGCGGCCCCCCGGTCACCACGCGGAGCCCAACCAGGCGATGGGCTTCTGTTTCTTCAACAACGTGGCCATCGCCGCCCGCGTGCTGCACACCAGGCTGGGGCTGCAGAGGATACTCATCGTCGACTGGGACGTGCATCACGGCAACGGCACCCAGCAGATATTCTACGAGGATCCTCACGTGCTGTACATGAGCATCCACCGGCACGACGACGGGAACTTCTTCCCTGGAACGGGAGCGGCCACCGAGTGCGGCGCCGGCCCGGGGCTCGGCTACAACGTGAACGTGCCCTGGAGCTGCGGCACCGACCCGCCCCTGGGGGACGCGGAGTACCTGGCCGCCTTCCGCTCCGTCATCATGCCCATCGCCAAGGAGTACGACCCCGAGATAGTGCTCGTGTCGTGCGGGTTCGACGCCGCGGCCGGGCACCCGGCGCCGCTCGGCGGGTACAGCGTGTCCGCCGCCTGCTTCGCGCACATGACCAGGGACCTCATGCAGCTCGCCAACGGCAAGGTGGTGATGTCGCTGGAGGGCGGCTACGACCTGGCGGCCATGTGCGACTGCGCGCAGGAGGTGGTCCGCGCGCTGCTGGGGGACCGCCCCCCCGCGCCCCCCTACGCCGAGCTGTGCCGCGCCCCCCACCCCCGCGCCCAGCACGCGCTCGCCGCCACGATTGCCGTGCAGGCGCAGCACTGGCCCACCATCAAGCGGTACAGCAGCCTGGTGGGCATGTCTGCGTTAGAGGCCGCGCCCTCGGTGGCGGCGGCCCGGCTCGGAGCCCTGCAGAGCGAACGGGACGCCGCCGACACCGCAGCCGCCATGGCGACGCTGTCAATGCACCAGCACAACCGGCCCGTGTCCACGTCTGCAGAAAGTTCAAGGTCGGTTTCGGAGGAGCCCATGGAGCAGGACGAAGCGAAAGATGACTATTTGTGTGACAACAAGGAATAA
Protein
MAHEGPARTEGSPHHTPSPPHLPHLPVTDSGFHHQIMQNHSPRKNHIVERAKQTLENSFLQQLKKQQQLQQEILLQHFQQQRQQLAREHENQMREHLKLWEQQKKAMEEAALREAREAREAREARERHERDRVELMRKKDIHEHSANASTQVKQKLQEFIKKKQANANGTVTTASYRNWGIVKSSSGESITSTGATATHPYRLTAPLPLNLNAPPPQPSPADYPLRKTASEPNMLKQRLKARVIERRASPLARRPLRTRHKHRNCEGGSPRGSPPGGCAAAPIREEEENGSRAAEFLFSSPSLPNISLGRPHVGPRLHPPPAAHTPAPVVLPPVSEAEALVVGGAGGRPVQRLGRTHSAPLPLGDPALQPPPVHHYLRDQIRKTVLTRAHDAAVQLREEEGEVIDLTSKRNDPPAPVEPAPLTRALSSPLVGARVPATGLAYDPLMLKHGCACGQHAPTHPEHGGRLQSVWARLCETGLAARTERTRPRKATLDELQSVHTEAHVQVFAGRGRDGAGVGAGGGAGGAVRQLVRLACGGLGVDSDTAWSDLHTAPAARLAAGALLDLALRTARGHLRNGFAVVRPPGHHAEPNQAMGFCFFNNVAIAARVLHTRLGLQRILIVDWDVHHGNGTQQIFYEDPHVLYMSIHRHDDGNFFPGTGAATECGAGPGLGYNVNVPWSCGTDPPLGDAEYLAAFRSVIMPIAKEYDPEIVLVSCGFDAAAGHPAPLGGYSVSAACFAHMTRDLMQLANGKVVMSLEGGYDLAAMCDCAQEVVRALLGDRPPAPPYAELCRAPHPRAQHALAATIAVQAQHWPTIKRYSSLVGMSALEAAPSVAAARLGALQSERDAADTAAAMATLSMHQHNRPVSTSAESSRSVSEEPMEQDEAKDDYLCDNKE
Summary
Description
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events.
Catalytic Activity
Hydrolysis of an N(6)-acetyl-lysine residue of a histone to yield a deacetylated histone.
Similarity
Belongs to the histone deacetylase family. HD type 2 subfamily.
Uniprot
EC Number
3.5.1.98
EMBL
Proteomes
Pfam
PF00850 Hist_deacetyl
Interpro
SUPFAM
SSF52768
SSF52768
Gene 3D
ProteinModelPortal
PDB
5ZOP
E-value=2.2373e-123,
Score=1135
Ontologies
Topology
Subcellular location
Nucleus
Length:
897
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0169
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00003
outside
1 - 897
Population Genetic Test Statistics
Pi
2.711671
Theta
23.881606
Tajima's D
-1.216707
CLR
1.632387
CSRT
0.101044947752612
Interpretation
Uncertain
