Pre Gene Modal
BGIBMGA009835
Annotation
PREDICTED:_structural_maintenance_of_chromosomes_protein_1A_[Papilio_xuthus]
Full name
Structural maintenance of chromosomes protein
Location in the cell
Nuclear Reliability : 2.574
Sequence
CDS
ATGCCTGCGTTTCTTAAATATATCGATATGGAAAATTTCAAGACCTATAGAGGACATCACCGCATAGGTCCTCTGAAGTCTTTTACCGCTGTTGTTGGCCCTAATGGATCAGGAAAGTCCAACTTCATGGACGCTGTGAGCTTTGTGATGGGTGAGAAGACATCATTGTTGCGTGTGAAGAGGTTGAGTGACTTGATCCATGGTGCTTCAATCAACAAACCTGTTTCTCGAAGTGCATCTGTTACGGCAACGTTTGTTCTGGAAGACATGACTGAGAAACATTTCCAGAGATCAGTGATTGGACAATCGTCTGAACATAAAATTGATGGCCAGTCTGTATCAGTAAGTAACTATCTTGGTGAACTAGAGAAATTGGGCATCAATGTGAAAGCCAAGAACTTCCTAGTCTTCCAGGGTGCCGTCGAGAGCATTGCCATGAAGAATCCTAAAGAAAGGACTACTCTGTTTGAGGAGATCAGCGGGTCTGGAGTGTTAAAGGAACAGTATGAAGCTTGCCGTGCTGAAGTAAACCGAGCTGATGAGGAGGCTCAATTCTCCTATCAGAAAAAGAAGGGAGTGGCAGCTGAGAGGAAAGAAGCAAAGTTTGAGAAGGAAGAAGCGGAGAAATACACAAGACTTAAGCAAGAATTGCAAGAGCAAAAAGTGGAACTTCAACTTTTCCATCTGTATCACAACGAGAGAGAGATCCAGGCTTATGAAGAAGATTTGCAACACAAACAACAAGAATTAGCAAAAATTGAGAAGAAAAGACAGAAGGCTGAAGAAGCATTAAAGGAAAAGAAAAAAGAAGCCGGAACCGTACAGAGAGAGCTCGCAAAGATTGAACAGGACATTAGGGAAGTGGAAGCAGAGATTTCAAAAAAACGACCAACTTTCATAAAAGCAAAAGAACGAGTCACTCACACCCAGAAGAAGCTTGAAAGTGCTCAAAAGACATTAGAACAAGCGCGGAAAGCTCATGAGGCCCATCAGGATGACATCAGGACTTTGGAGGAGGAGCTTAGAACCTTGGAGCAGCAAAAGGCAACATGGGAAACAGCTAGTGGTACCGGTCACAGTGGGAAGGCTGATGTACATTTGGAGGAGGCCCAGATCCGTGAATATGAAGAGCTGAAGATGGAAGCATCCCGTCAGGCAGCTCGCTACCTCCAGGAACTGGATTCCGTGAATAGGGAACAGAAGGCAGACCAGGATCGTCTGGACAATGAGCTGAGGAAGAAGGGAGAGCTGGAAAACAAGCATCGGCAGAAGGGGCACGAGAGAAACGAAGCTGTGAAGAGAGTGGACAAACTAAATGAACACATCAAGAGTTCGGAACAGGCCCTTGAGGAACAGAGGAGACTACGAGCTGAATTACAGGCGGACGTGGGTTCGTGCCGCGGGCGGGCCGCCAGCCTGCAGACGCAGCTGGAGGACGTGGCGGCGCAGCTGGGCGACGCGCGCGTGGACAAGCACGAGGAGGCGCGCCGCAGGAAGAAGCAGGAGATCGTCGAGAGCTTCAAGCGAGACATCCCCGGGGTTTACGACAGAATGATCAACATGTGCCAGCCGACCCACAAAAGGTACAACGTGGCCATCACCAAGGTCCTCGGGAAGTACATGGAAGCTATCGTGGTGGACACCGAGAAGACAGCCCGGAGGTGTATCCAGGTGCTGAAAGAAAGAATGCTGGAACCGGAGACTTTCCTCCCCCTGGACTACATACAGGCCAAACCGCTGCGGGAGAGACTGAGAGACATAAAGGAGCCCAAGAACGTGAAGCTGCTGTTCGACGTGCTCCGCTTCGAGCCGGCCGCCATCCACCGCGCCGTGCTCTTCGTCACCAACAACGCGCTCGTGTGCGAGACCCCCGAGGACGCGTCGCGCGTCGCCTACGACCTCGACCGGAACAAGAACAGCAGATACGATGCGCTGGCGCTGGACGGCACGTTCTACCAGAAGTCTGGCATCATATCGGGGGGCTCGTTGGATCTGGCGAGGAAAGCTAAACGATGGGACGAGAAGCATCTTTCACAGCTTAAAGCTAAAAAGGAGAAGCTAACAGAAGAGCTCCGGGAGTCGATGAAGAAGTCCCGCAAGGAGTCGGAGCTGACCACCGTAGACTCGCAGATCCGGGGGCTCGAGTCCAGACTCAAGTACGCCGTCACCGACAGGGACACCACCTTAAAACAAATTAAGACGTTGGATGCTGAGATCGTTGAGCTGGAGAGAAAGATGGAAACTTTTGGGCCTCAGATCGAGGAGATCGAGCGCACCATCCGGCTGCGCGACGCCAAGATCCAGGAGGTGAAGGAGAACATGAACAACGTGGAGGACGTCGTGTTCAAGGCGTTCTGTCGGGACATCGGCGTCGCCAACATCCGACAGTACGAGGAGCGCGAGCTGCGCGCCCAGCAGGAGCGCGCCAAGCGGCGGATGGAGTTTGAAGCGCAAATCGACAGGGTCGCCTCCAACCTGGAGTTCGAGCGATCCCGGGACACGCAGAAGAACGTGACGCGGTGGGAGCGCGCGGTGCAGGACGCCGAGGACGAGCTGGAGGGCGGGCGCCAGGCGGAGGCCAAGCAGCGCGCCGACATCGACCACGAGCTGCGCCGCGCCGACACGCTCAAGGCGGACCGGGCCGCCGCGCGCACGCACCTCGAGAAGGCGGAGGAGGACGTCAACAAGGCGCGGAAAGAGGTGTCCAGCATCCAGAAGGACATCCAGAGCGTCCAGAAGCAGATGGCCAGCATCGAGGCGCGCATCGAGAGCAAGCGCAGCGAGCGACACAACATACTGCGGCAGTGCAAGATCGATGACATAATAATACCCTTATTGGAGGGTAGCCTAGACGACACGGCCGACACGGAGTCCGACCCCTCCTCCATGTCCACCACGCAGCAGTACAGGAAGGAATCCAGGATTCGTGTGGACTACAGCATGCTGTCGGACAGCCTGCGCGACCTGGAGGAGGCCGACGAGGTCCGGCGCCGCGCGGACAAGCTGCAGAAGGCGATCAACTCCCTGCAGACGACCGTCGACAAGATCCAGGCGCCCAACATGCGGGCCATGCAGAAGCTGACGGAGGTGCGGGAGAAGGTGAACGCTACCAACGAGGCCTTCGTGGCGGCGAGGAAGCGCGCGCACCGCGCCAAGCTCGCCTTCGAGAAGGTGAAGAAGGAGCGCCACGACAAGTTCATGGACTGCTTCGAGCACGTCGCCAACGAGATCGACGCCATCTACAAGGCGCTGGCGATGAACCAGTCGGCGCAGGCGTTCCTGGGTCCCGAGAACCCCGAGGAGCCGTACCTGGACGGCATCAACTACAACTGCGTGGCGCCCGGGAAGCGCTTCCAGCCCATGTCCAACCTCTCCGGCGGGGAGAAGACGGTGGCCGCGCTAGCGCTGCTGTTCGCCATCCACAGCTACCAGCCGGCGCCGTTCTTTCTGCTGGACGAGATAGACGCGGCCCTGGACAACACCAACATAGGCAAGGTCGCCTCCTACATCCGCTCCAAGAAGGGCTGTCTGCAGACCATCGTCATCTCGCTCAAGGAGGAGTTCTATGGGTGCGCCGATGCGCTGGTCGGCATCTGCTCGGAGCCGGCGGACTGCCTCGTGAGTGACGTCATCACCCTCAGCCTGGAGAACTACGCGGATTAG
Protein
MPAFLKYIDMENFKTYRGHHRIGPLKSFTAVVGPNGSGKSNFMDAVSFVMGEKTSLLRVKRLSDLIHGASINKPVSRSASVTATFVLEDMTEKHFQRSVIGQSSEHKIDGQSVSVSNYLGELEKLGINVKAKNFLVFQGAVESIAMKNPKERTTLFEEISGSGVLKEQYEACRAEVNRADEEAQFSYQKKKGVAAERKEAKFEKEEAEKYTRLKQELQEQKVELQLFHLYHNEREIQAYEEDLQHKQQELAKIEKKRQKAEEALKEKKKEAGTVQRELAKIEQDIREVEAEISKKRPTFIKAKERVTHTQKKLESAQKTLEQARKAHEAHQDDIRTLEEELRTLEQQKATWETASGTGHSGKADVHLEEAQIREYEELKMEASRQAARYLQELDSVNREQKADQDRLDNELRKKGELENKHRQKGHERNEAVKRVDKLNEHIKSSEQALEEQRRLRAELQADVGSCRGRAASLQTQLEDVAAQLGDARVDKHEEARRRKKQEIVESFKRDIPGVYDRMINMCQPTHKRYNVAITKVLGKYMEAIVVDTEKTARRCIQVLKERMLEPETFLPLDYIQAKPLRERLRDIKEPKNVKLLFDVLRFEPAAIHRAVLFVTNNALVCETPEDASRVAYDLDRNKNSRYDALALDGTFYQKSGIISGGSLDLARKAKRWDEKHLSQLKAKKEKLTEELRESMKKSRKESELTTVDSQIRGLESRLKYAVTDRDTTLKQIKTLDAEIVELERKMETFGPQIEEIERTIRLRDAKIQEVKENMNNVEDVVFKAFCRDIGVANIRQYEERELRAQQERAKRRMEFEAQIDRVASNLEFERSRDTQKNVTRWERAVQDAEDELEGGRQAEAKQRADIDHELRRADTLKADRAAARTHLEKAEEDVNKARKEVSSIQKDIQSVQKQMASIEARIESKRSERHNILRQCKIDDIIIPLLEGSLDDTADTESDPSSMSTTQQYRKESRIRVDYSMLSDSLRDLEEADEVRRRADKLQKAINSLQTTVDKIQAPNMRAMQKLTEVREKVNATNEAFVAARKRAHRAKLAFEKVKKERHDKFMDCFEHVANEIDAIYKALAMNQSAQAFLGPENPEEPYLDGINYNCVAPGKRFQPMSNLSGGEKTVAALALLFAIHSYQPAPFFLLDEIDAALDNTNIGKVASYIRSKKGCLQTIVISLKEEFYGCADALVGICSEPADCLVSDVITLSLENYAD
Summary
Similarity
Belongs to the SMC family.
Uniprot
H9JJY4
A0A2H1WK95
A0A194PS52
A0A2A4K815
S4PM86
A0A212FGH0
+ More
A0A194RCU8 A0A2J7QPR3 A0A067RHM4 E2B6C3 D6WWY2 A0A1Y1MWJ6 A0A291S6X4 A0A026VXY9 E2ABT2 A0A0N0BGL5 A0A1W4XLN4 A0A2A3EF39 A0A087ZYL5 F4W8Y1 A0A158NP42 A0A151ILC7 A0A232FFB6 A0A195F0V4 A0A0P4VW93 A0A195DWT6 A0A1B6LYD5 J9K5B6 A0A2H8TVT0 A0A2S2QIB6 A0A2P2I4S7 A0A164WFH0 A0A0N8DB39 A0A0A9YLV3 A0A0P5SIA5 E9GJ33 A0A182J865 E0VVX1 A0A182H986 U4UN83 U5EXJ4 Q8I954 A0A1S4FBX8 Q178Q7 A0A2M4A9V7 A0A2M4BA64 A0A1B6DY45 A0A0L7QJS8 A0A182M1K9 A0A069DXE1 B0WY22 Q7QD42 A0A1Q3F7A2 A0A2M4CUI9 A0A336MXJ1 A0A182UWW2 A0A0V0G5N2 A0A1J1J8I8 A0A3F2YTY7 A0A182Q7V9 A0A1A9UFQ4 W8C8Y2 A0A1A9XRG2 A0A1B0BVV1 A0A1A9ZA09 A0A034W888 A0A0K8UST1 T1P898 T1I0K2 A0A1I8PIF5 A0A1B0FIR3 B3MST3 A0A0A1WHM9 A0A1B0D6M0 A0A0L0BMS7 A0A3B0JK50 Q9VCD8 A0A182LG27 Q298K8 A0A1W4V5M0 A0A0K8UKN7 Q9N6I4 B3P7A0 B4PL25 B4NBN8 B4MBZ5 B4G429 B4K7D0 A0A0M4EQU7 B4JIH2 N6TQU0 A0A0P5YC08 A0A151X8F7 A0A0K2U3J1 A0A131YK12 A0A131XJX2 L7M269 A0A224Z568 A0A131YDX8 A0A224Z9C4
A0A194RCU8 A0A2J7QPR3 A0A067RHM4 E2B6C3 D6WWY2 A0A1Y1MWJ6 A0A291S6X4 A0A026VXY9 E2ABT2 A0A0N0BGL5 A0A1W4XLN4 A0A2A3EF39 A0A087ZYL5 F4W8Y1 A0A158NP42 A0A151ILC7 A0A232FFB6 A0A195F0V4 A0A0P4VW93 A0A195DWT6 A0A1B6LYD5 J9K5B6 A0A2H8TVT0 A0A2S2QIB6 A0A2P2I4S7 A0A164WFH0 A0A0N8DB39 A0A0A9YLV3 A0A0P5SIA5 E9GJ33 A0A182J865 E0VVX1 A0A182H986 U4UN83 U5EXJ4 Q8I954 A0A1S4FBX8 Q178Q7 A0A2M4A9V7 A0A2M4BA64 A0A1B6DY45 A0A0L7QJS8 A0A182M1K9 A0A069DXE1 B0WY22 Q7QD42 A0A1Q3F7A2 A0A2M4CUI9 A0A336MXJ1 A0A182UWW2 A0A0V0G5N2 A0A1J1J8I8 A0A3F2YTY7 A0A182Q7V9 A0A1A9UFQ4 W8C8Y2 A0A1A9XRG2 A0A1B0BVV1 A0A1A9ZA09 A0A034W888 A0A0K8UST1 T1P898 T1I0K2 A0A1I8PIF5 A0A1B0FIR3 B3MST3 A0A0A1WHM9 A0A1B0D6M0 A0A0L0BMS7 A0A3B0JK50 Q9VCD8 A0A182LG27 Q298K8 A0A1W4V5M0 A0A0K8UKN7 Q9N6I4 B3P7A0 B4PL25 B4NBN8 B4MBZ5 B4G429 B4K7D0 A0A0M4EQU7 B4JIH2 N6TQU0 A0A0P5YC08 A0A151X8F7 A0A0K2U3J1 A0A131YK12 A0A131XJX2 L7M269 A0A224Z568 A0A131YDX8 A0A224Z9C4
Pubmed
19121390
26354079
23622113
22118469
24845553
20798317
+ More
18362917 19820115 28004739 28992199 24508170 30249741 21719571 21347285 28648823 25401762 26823975 21292972 20566863 26483478 23537049 14660695 17510324 26334808 12364791 14747013 17210077 24495485 25348373 25315136 17994087 25830018 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 15632085 10806064 17550304 26830274 28049606 25576852 28797301
18362917 19820115 28004739 28992199 24508170 30249741 21719571 21347285 28648823 25401762 26823975 21292972 20566863 26483478 23537049 14660695 17510324 26334808 12364791 14747013 17210077 24495485 25348373 25315136 17994087 25830018 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 15632085 10806064 17550304 26830274 28049606 25576852 28797301
EMBL
BABH01026721
BABH01026722
ODYU01009232
SOQ53503.1
KQ459595
KPI95793.1
+ More
NWSH01000074 PCG79933.1 GAIX01003710 JAA88850.1 AGBW02008669 OWR52835.1 KQ460367 KPJ15447.1 NEVH01012088 PNF30565.1 KK852678 KDR18667.1 GL445930 EFN88816.1 KQ971361 EFA08076.1 GEZM01019965 JAV89508.1 MF433016 ATL75367.1 KK107648 QOIP01000008 EZA48341.1 RLU19352.1 GL438356 EFN69109.1 KQ435775 KOX74954.1 KZ288262 PBC30375.1 GL887974 EGI69340.1 ADTU01022022 ADTU01022023 KQ977120 KYN05557.1 NNAY01000289 OXU29451.1 KQ981891 KYN33787.1 GDRN01103626 GDRN01103625 JAI58049.1 KQ980167 KYN17370.1 GEBQ01011284 JAT28693.1 ABLF02028753 ABLF02028757 GFXV01006551 MBW18356.1 GGMS01008271 MBY77474.1 IACF01003421 LAB69037.1 LRGB01001253 KZS13214.1 GDIP01051135 GDIQ01002820 JAM52580.1 JAN91917.1 GBHO01009557 GDHC01006499 JAG34047.1 JAQ12130.1 GDIP01139427 JAL64287.1 GL732547 EFX80555.1 DS235815 EEB17527.1 JXUM01030067 JXUM01030068 JXUM01030069 JXUM01030070 KQ560872 KXJ80531.1 KB632395 ERL94587.1 GANO01000922 JAB58949.1 AJ535203 CAD59403.1 CH477360 EAT42684.1 GGFK01004077 MBW37398.1 GGFJ01000804 MBW49945.1 GEDC01006700 JAS30598.1 KQ415041 KOC58863.1 AXCM01001293 GBGD01000156 JAC88733.1 DS232179 EDS36832.1 AAAB01008859 EAA07609.3 GFDL01011601 JAV23444.1 GGFL01004771 MBW68949.1 UFQT01001447 SSX30608.1 GECL01002748 JAP03376.1 CVRI01000070 CRL07225.1 AXCN02001200 GAMC01002634 JAC03922.1 JXJN01021483 GAKP01008028 JAC50924.1 GDHF01022592 JAI29722.1 KA644824 AFP59453.1 ACPB03018149 CCAG010023921 CH902623 EDV30323.1 GBXI01015945 JAC98346.1 AJVK01012086 AJVK01012087 AJVK01012088 JRES01001712 KNC20569.1 OUUW01000007 SPP82724.1 AE014297 AY118636 AAF56231.1 AAM50005.1 CM000070 EAL27947.2 GDHF01025077 JAI27237.1 AF225909 AJ271845 AAF43149.1 CAB76376.1 CH954182 EDV53920.1 CM000160 EDW99010.1 CH964232 EDW81202.1 CH940656 EDW58616.1 CH479179 EDW24440.1 CH933806 EDW14254.1 CP012526 ALC47438.1 CH916369 EDV93053.1 APGK01024336 APGK01024337 KB740543 ENN80428.1 GDIP01061285 JAM42430.1 KQ982422 KYQ56614.1 HACA01015478 CDW32839.1 GEDV01009715 JAP78842.1 GEFH01002151 JAP66430.1 GACK01006909 JAA58125.1 GFPF01012093 MAA23239.1 GEDV01012291 JAP76266.1 GFPF01012094 MAA23240.1
NWSH01000074 PCG79933.1 GAIX01003710 JAA88850.1 AGBW02008669 OWR52835.1 KQ460367 KPJ15447.1 NEVH01012088 PNF30565.1 KK852678 KDR18667.1 GL445930 EFN88816.1 KQ971361 EFA08076.1 GEZM01019965 JAV89508.1 MF433016 ATL75367.1 KK107648 QOIP01000008 EZA48341.1 RLU19352.1 GL438356 EFN69109.1 KQ435775 KOX74954.1 KZ288262 PBC30375.1 GL887974 EGI69340.1 ADTU01022022 ADTU01022023 KQ977120 KYN05557.1 NNAY01000289 OXU29451.1 KQ981891 KYN33787.1 GDRN01103626 GDRN01103625 JAI58049.1 KQ980167 KYN17370.1 GEBQ01011284 JAT28693.1 ABLF02028753 ABLF02028757 GFXV01006551 MBW18356.1 GGMS01008271 MBY77474.1 IACF01003421 LAB69037.1 LRGB01001253 KZS13214.1 GDIP01051135 GDIQ01002820 JAM52580.1 JAN91917.1 GBHO01009557 GDHC01006499 JAG34047.1 JAQ12130.1 GDIP01139427 JAL64287.1 GL732547 EFX80555.1 DS235815 EEB17527.1 JXUM01030067 JXUM01030068 JXUM01030069 JXUM01030070 KQ560872 KXJ80531.1 KB632395 ERL94587.1 GANO01000922 JAB58949.1 AJ535203 CAD59403.1 CH477360 EAT42684.1 GGFK01004077 MBW37398.1 GGFJ01000804 MBW49945.1 GEDC01006700 JAS30598.1 KQ415041 KOC58863.1 AXCM01001293 GBGD01000156 JAC88733.1 DS232179 EDS36832.1 AAAB01008859 EAA07609.3 GFDL01011601 JAV23444.1 GGFL01004771 MBW68949.1 UFQT01001447 SSX30608.1 GECL01002748 JAP03376.1 CVRI01000070 CRL07225.1 AXCN02001200 GAMC01002634 JAC03922.1 JXJN01021483 GAKP01008028 JAC50924.1 GDHF01022592 JAI29722.1 KA644824 AFP59453.1 ACPB03018149 CCAG010023921 CH902623 EDV30323.1 GBXI01015945 JAC98346.1 AJVK01012086 AJVK01012087 AJVK01012088 JRES01001712 KNC20569.1 OUUW01000007 SPP82724.1 AE014297 AY118636 AAF56231.1 AAM50005.1 CM000070 EAL27947.2 GDHF01025077 JAI27237.1 AF225909 AJ271845 AAF43149.1 CAB76376.1 CH954182 EDV53920.1 CM000160 EDW99010.1 CH964232 EDW81202.1 CH940656 EDW58616.1 CH479179 EDW24440.1 CH933806 EDW14254.1 CP012526 ALC47438.1 CH916369 EDV93053.1 APGK01024336 APGK01024337 KB740543 ENN80428.1 GDIP01061285 JAM42430.1 KQ982422 KYQ56614.1 HACA01015478 CDW32839.1 GEDV01009715 JAP78842.1 GEFH01002151 JAP66430.1 GACK01006909 JAA58125.1 GFPF01012093 MAA23239.1 GEDV01012291 JAP76266.1 GFPF01012094 MAA23240.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000053240
UP000235965
+ More
UP000027135 UP000008237 UP000007266 UP000053097 UP000279307 UP000000311 UP000053105 UP000192223 UP000242457 UP000005203 UP000007755 UP000005205 UP000078542 UP000215335 UP000078541 UP000078492 UP000007819 UP000076858 UP000000305 UP000075880 UP000009046 UP000069940 UP000249989 UP000030742 UP000008820 UP000053825 UP000075883 UP000002320 UP000007062 UP000075903 UP000183832 UP000075881 UP000075886 UP000078200 UP000092443 UP000092460 UP000092445 UP000095301 UP000015103 UP000095300 UP000092444 UP000007801 UP000092462 UP000037069 UP000268350 UP000000803 UP000075882 UP000001819 UP000192221 UP000008711 UP000002282 UP000007798 UP000008792 UP000008744 UP000009192 UP000092553 UP000001070 UP000019118 UP000075809
UP000027135 UP000008237 UP000007266 UP000053097 UP000279307 UP000000311 UP000053105 UP000192223 UP000242457 UP000005203 UP000007755 UP000005205 UP000078542 UP000215335 UP000078541 UP000078492 UP000007819 UP000076858 UP000000305 UP000075880 UP000009046 UP000069940 UP000249989 UP000030742 UP000008820 UP000053825 UP000075883 UP000002320 UP000007062 UP000075903 UP000183832 UP000075881 UP000075886 UP000078200 UP000092443 UP000092460 UP000092445 UP000095301 UP000015103 UP000095300 UP000092444 UP000007801 UP000092462 UP000037069 UP000268350 UP000000803 UP000075882 UP000001819 UP000192221 UP000008711 UP000002282 UP000007798 UP000008792 UP000008744 UP000009192 UP000092553 UP000001070 UP000019118 UP000075809
Interpro
ProteinModelPortal
H9JJY4
A0A2H1WK95
A0A194PS52
A0A2A4K815
S4PM86
A0A212FGH0
+ More
A0A194RCU8 A0A2J7QPR3 A0A067RHM4 E2B6C3 D6WWY2 A0A1Y1MWJ6 A0A291S6X4 A0A026VXY9 E2ABT2 A0A0N0BGL5 A0A1W4XLN4 A0A2A3EF39 A0A087ZYL5 F4W8Y1 A0A158NP42 A0A151ILC7 A0A232FFB6 A0A195F0V4 A0A0P4VW93 A0A195DWT6 A0A1B6LYD5 J9K5B6 A0A2H8TVT0 A0A2S2QIB6 A0A2P2I4S7 A0A164WFH0 A0A0N8DB39 A0A0A9YLV3 A0A0P5SIA5 E9GJ33 A0A182J865 E0VVX1 A0A182H986 U4UN83 U5EXJ4 Q8I954 A0A1S4FBX8 Q178Q7 A0A2M4A9V7 A0A2M4BA64 A0A1B6DY45 A0A0L7QJS8 A0A182M1K9 A0A069DXE1 B0WY22 Q7QD42 A0A1Q3F7A2 A0A2M4CUI9 A0A336MXJ1 A0A182UWW2 A0A0V0G5N2 A0A1J1J8I8 A0A3F2YTY7 A0A182Q7V9 A0A1A9UFQ4 W8C8Y2 A0A1A9XRG2 A0A1B0BVV1 A0A1A9ZA09 A0A034W888 A0A0K8UST1 T1P898 T1I0K2 A0A1I8PIF5 A0A1B0FIR3 B3MST3 A0A0A1WHM9 A0A1B0D6M0 A0A0L0BMS7 A0A3B0JK50 Q9VCD8 A0A182LG27 Q298K8 A0A1W4V5M0 A0A0K8UKN7 Q9N6I4 B3P7A0 B4PL25 B4NBN8 B4MBZ5 B4G429 B4K7D0 A0A0M4EQU7 B4JIH2 N6TQU0 A0A0P5YC08 A0A151X8F7 A0A0K2U3J1 A0A131YK12 A0A131XJX2 L7M269 A0A224Z568 A0A131YDX8 A0A224Z9C4
A0A194RCU8 A0A2J7QPR3 A0A067RHM4 E2B6C3 D6WWY2 A0A1Y1MWJ6 A0A291S6X4 A0A026VXY9 E2ABT2 A0A0N0BGL5 A0A1W4XLN4 A0A2A3EF39 A0A087ZYL5 F4W8Y1 A0A158NP42 A0A151ILC7 A0A232FFB6 A0A195F0V4 A0A0P4VW93 A0A195DWT6 A0A1B6LYD5 J9K5B6 A0A2H8TVT0 A0A2S2QIB6 A0A2P2I4S7 A0A164WFH0 A0A0N8DB39 A0A0A9YLV3 A0A0P5SIA5 E9GJ33 A0A182J865 E0VVX1 A0A182H986 U4UN83 U5EXJ4 Q8I954 A0A1S4FBX8 Q178Q7 A0A2M4A9V7 A0A2M4BA64 A0A1B6DY45 A0A0L7QJS8 A0A182M1K9 A0A069DXE1 B0WY22 Q7QD42 A0A1Q3F7A2 A0A2M4CUI9 A0A336MXJ1 A0A182UWW2 A0A0V0G5N2 A0A1J1J8I8 A0A3F2YTY7 A0A182Q7V9 A0A1A9UFQ4 W8C8Y2 A0A1A9XRG2 A0A1B0BVV1 A0A1A9ZA09 A0A034W888 A0A0K8UST1 T1P898 T1I0K2 A0A1I8PIF5 A0A1B0FIR3 B3MST3 A0A0A1WHM9 A0A1B0D6M0 A0A0L0BMS7 A0A3B0JK50 Q9VCD8 A0A182LG27 Q298K8 A0A1W4V5M0 A0A0K8UKN7 Q9N6I4 B3P7A0 B4PL25 B4NBN8 B4MBZ5 B4G429 B4K7D0 A0A0M4EQU7 B4JIH2 N6TQU0 A0A0P5YC08 A0A151X8F7 A0A0K2U3J1 A0A131YK12 A0A131XJX2 L7M269 A0A224Z568 A0A131YDX8 A0A224Z9C4
PDB
2WD5
E-value=1.77136e-51,
Score=517
Ontologies
GO
PANTHER
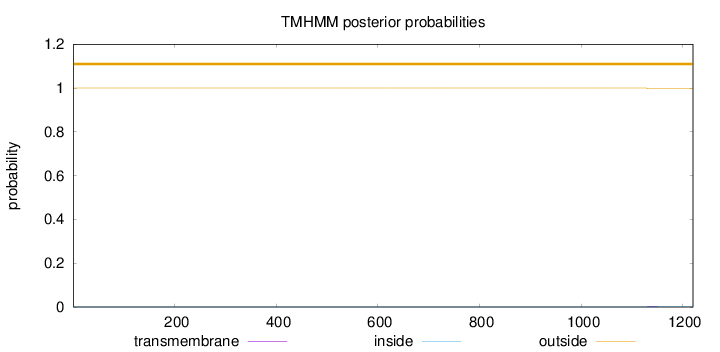
Topology
Subcellular location
Nucleus
Length:
1220
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03693
Exp number, first 60 AAs:
0.00119
Total prob of N-in:
0.00007
outside
1 - 1220
Population Genetic Test Statistics
Pi
24.045292
Theta
22.448143
Tajima's D
0.594628
CLR
1.688965
CSRT
0.546322683865807
Interpretation
Uncertain
