Gene
KWMTBOMO04809
Pre Gene Modal
BGIBMGA009914
Annotation
PREDICTED:_uncharacterized_protein_LOC106133170_isoform_X2_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.215 Nuclear Reliability : 1.081
Sequence
CDS
ATGTCGTTCCGGAGGCTGATGAGCGCTGTGCGATCTGCGCGGGAAGCCATAGCCAACAGCGCGCCCCTGGCGGACCGACTCCTCAAATCGCTGCTTTCCGTCGTGGTGATACTGTGCATAGCGGTGTCCTCTCCACCGCCGGCTTCCGCGAAGCCTTTTGCTATAAGCTTCCCGGCAGGCTGGTCCCTTGCCGGCTTCGTGGGCAGTGTGGGCGCTCGAGCGGATGAACCCCAGCGGAAGGAGCCGGCGGCAGGCGTGAAGCCTTACACGGGCCGACGAATCAGCAACGATACATTGAGAATGATATACTACCACGACCAGACCGTGGCGGTCGTGGAGCTAGGAACGAACAAGTTATTGCTCAACTGCGAACTCATCGAGACATACGATGAAGAAGATACGAAGAGACTGCTCCGTCAGTTGAGCTCCATCAACCGTCCTCTGGAAGTCACCTTCCAGCAAATGGTGAATCTAATGGCACAATGCCAACAGGTAGAAGGTGTAGAAGCTCAAGACGAGACGGGACGAGAGTTCAATGGCTGGAGAGAAAAGAGTAGCAGTGCAGCGGCTAAAGCGCGGCTGGACGCAGGAGGAGCACACGCAGGGCTCCTGGGCGGCAGTCCGCTCACACTCCTCCAGGGCATCATCCCTGGTACAAAATGGTGCGGTACTGGGGATATAGCCGCCGATTACCATGACTTGGGCGCTGACAGGCGACTGGACCGCTGCTGTAGAACCCATGACCTTTGCCCAACTAAAGTTAGAGCATTCTCCAAACGCTACAACCTTATTAATAATTCTTTATACAGCAAATCGCATTGCGCATGCGACGACATGTTATTCGACTGTTTGAAAGCCACTAATACATCCGCCTCACATCTAATGGGCCATATTTATTTTAACCTAGTTCAAGTCCCGTGCTTAGAAGACATTCCAAACGGAAGACGGTTTAGGAACGCGAAAGAAGGTTTTTGA
Protein
MSFRRLMSAVRSAREAIANSAPLADRLLKSLLSVVVILCIAVSSPPPASAKPFAISFPAGWSLAGFVGSVGARADEPQRKEPAAGVKPYTGRRISNDTLRMIYYHDQTVAVVELGTNKLLLNCELIETYDEEDTKRLLRQLSSINRPLEVTFQQMVNLMAQCQQVEGVEAQDETGREFNGWREKSSSAAAKARLDAGGAHAGLLGGSPLTLLQGIIPGTKWCGTGDIAADYHDLGADRRLDRCCRTHDLCPTKVRAFSKRYNLINNSLYSKSHCACDDMLFDCLKATNTSASHLMGHIYFNLVQVPCLEDIPNGRRFRNAKEGF
Summary
Similarity
Belongs to the phospholipase A2 family.
Uniprot
A0A2A4K7E0
A0A212FHD2
U5EP92
A0A1J1J0M9
A0A336LL42
A0A182PL18
+ More
C0LTQ2 A0A1S4FNL5 Q16UK2 A0A182R7V4 T1PJK7 W5JW74 A0A182HMV0 A0A182UDD1 A0A182VPD7 A0A3B0J7J1 A0A0L0CQY4 A0A182XNL1 B4NQ77 A0A1Y1K121 B4M833 A0A2M4AG40 Q8SZ42 A0A182Y4V6 A0A182FMF6 B0XJN6 X2JBI9 A0A2M4AI58 B3MYZ1 B3NWS6 Q9VYG9 B4IGM6 A0A0K8UIM1 Q29H67 B4R418 A0A034VNU6 A0A1A9WAZ0 B4L6F9 A0A1B0AJ05 A0A1A9UHU7 A0A0A1WHB1 A0A1W4V7T5 B4Q211 B4JKL4 W8AUB2 A0A1I8NLM2 A0A1L8DPX4 B4H084 V5K5P8 Q2TJG0 V5K6B7 Q0ZS49 A0A1W4WFV5 C6FFT9 A0A1B0CWT6 A0A1B0FM16 J9JV77 A0A2H8TEH8 A0A2S2N722 A0A1B6DBG7 A0A0N0BF21 A0A154PT88 A0A067QSN2 A0A3L8DTG3 A0A0C9R8Q7 A0A2A3ENE0 A0A1S3D1V7 A0A1B6K949 A0A1B6I8W4 A0A026X0W9 A0A195BQR4 A0A158NDW9 E9IGS3 F4W7U7 A0A0J7L2K4 B0XJ99
C0LTQ2 A0A1S4FNL5 Q16UK2 A0A182R7V4 T1PJK7 W5JW74 A0A182HMV0 A0A182UDD1 A0A182VPD7 A0A3B0J7J1 A0A0L0CQY4 A0A182XNL1 B4NQ77 A0A1Y1K121 B4M833 A0A2M4AG40 Q8SZ42 A0A182Y4V6 A0A182FMF6 B0XJN6 X2JBI9 A0A2M4AI58 B3MYZ1 B3NWS6 Q9VYG9 B4IGM6 A0A0K8UIM1 Q29H67 B4R418 A0A034VNU6 A0A1A9WAZ0 B4L6F9 A0A1B0AJ05 A0A1A9UHU7 A0A0A1WHB1 A0A1W4V7T5 B4Q211 B4JKL4 W8AUB2 A0A1I8NLM2 A0A1L8DPX4 B4H084 V5K5P8 Q2TJG0 V5K6B7 Q0ZS49 A0A1W4WFV5 C6FFT9 A0A1B0CWT6 A0A1B0FM16 J9JV77 A0A2H8TEH8 A0A2S2N722 A0A1B6DBG7 A0A0N0BF21 A0A154PT88 A0A067QSN2 A0A3L8DTG3 A0A0C9R8Q7 A0A2A3ENE0 A0A1S3D1V7 A0A1B6K949 A0A1B6I8W4 A0A026X0W9 A0A195BQR4 A0A158NDW9 E9IGS3 F4W7U7 A0A0J7L2K4 B0XJ99
Pubmed
22118469
18362917
19931277
19820115
17510324
25315136
+ More
20920257 23761445 26108605 17994087 28004739 18057021 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 25348373 25830018 17550304 24495485 24587463 16154670 16539713 19555500 24845553 30249741 24508170 21347285 21282665 21719571
20920257 23761445 26108605 17994087 28004739 18057021 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 25348373 25830018 17550304 24495485 24587463 16154670 16539713 19555500 24845553 30249741 24508170 21347285 21282665 21719571
EMBL
NWSH01000074
PCG79936.1
AGBW02008504
OWR53152.1
GANO01004808
JAB55063.1
+ More
CVRI01000063 CRL04401.1 UFQT01000041 SSX18690.1 FJ768719 KQ971361 ACN42748.1 EFA07968.1 CH477620 EAT38212.1 KA648879 AFP63508.1 ADMH02000109 ETN67803.1 APCN01004913 OUUW01000003 SPP77775.1 JRES01000037 KNC34661.1 CH964291 EDW86302.1 GEZM01096056 JAV55179.1 CH940653 EDW62309.1 KRF80553.1 GGFK01006267 MBW39588.1 AY071131 AAL48753.1 DS233566 EDS30555.1 AE014298 AHN59655.1 GGFK01007140 MBW40461.1 CH902632 EDV32835.1 KPU74162.1 KPU74163.1 CH954180 EDV47238.1 AAF48228.2 CH480836 EDW48976.1 GDHF01025926 JAI26388.1 CH379064 EAL31891.2 CM000366 EDX17785.1 GAKP01014813 GAKP01014809 GAKP01014805 JAC44139.1 CH933812 EDW05955.2 GBXI01016412 JAC97879.1 CM000162 EDX01532.2 CH916370 EDW00117.1 GAMC01018257 GAMC01018256 GAMC01018255 JAB88298.1 GFDF01005572 JAV08512.1 CH479200 EDW29679.1 KC170982 AGT96476.1 AY861671 AAX54852.1 KC170949 AGT96443.1 DQ154097 ABA43062.1 EZ000627 ACS93491.1 AJWK01032852 AJWK01032853 AJWK01032854 CCAG010015359 ABLF02021831 GFXV01000701 MBW12506.1 GGMR01000259 MBY12878.1 GEDC01014316 JAS22982.1 KQ435821 KOX72448.1 KQ435078 KZC14498.1 KK853603 KDR06363.1 QOIP01000004 RLU23674.1 GBYB01012744 JAG82511.1 KZ288215 PBC32561.1 GEBQ01032004 JAT07973.1 GECU01024335 JAS83371.1 KK107046 EZA61723.1 KQ976424 KYM88478.1 ADTU01013057 GL763054 EFZ20207.1 GL887888 EGI69617.1 LBMM01001117 KMQ96893.1 DS233465 EDS30114.1
CVRI01000063 CRL04401.1 UFQT01000041 SSX18690.1 FJ768719 KQ971361 ACN42748.1 EFA07968.1 CH477620 EAT38212.1 KA648879 AFP63508.1 ADMH02000109 ETN67803.1 APCN01004913 OUUW01000003 SPP77775.1 JRES01000037 KNC34661.1 CH964291 EDW86302.1 GEZM01096056 JAV55179.1 CH940653 EDW62309.1 KRF80553.1 GGFK01006267 MBW39588.1 AY071131 AAL48753.1 DS233566 EDS30555.1 AE014298 AHN59655.1 GGFK01007140 MBW40461.1 CH902632 EDV32835.1 KPU74162.1 KPU74163.1 CH954180 EDV47238.1 AAF48228.2 CH480836 EDW48976.1 GDHF01025926 JAI26388.1 CH379064 EAL31891.2 CM000366 EDX17785.1 GAKP01014813 GAKP01014809 GAKP01014805 JAC44139.1 CH933812 EDW05955.2 GBXI01016412 JAC97879.1 CM000162 EDX01532.2 CH916370 EDW00117.1 GAMC01018257 GAMC01018256 GAMC01018255 JAB88298.1 GFDF01005572 JAV08512.1 CH479200 EDW29679.1 KC170982 AGT96476.1 AY861671 AAX54852.1 KC170949 AGT96443.1 DQ154097 ABA43062.1 EZ000627 ACS93491.1 AJWK01032852 AJWK01032853 AJWK01032854 CCAG010015359 ABLF02021831 GFXV01000701 MBW12506.1 GGMR01000259 MBY12878.1 GEDC01014316 JAS22982.1 KQ435821 KOX72448.1 KQ435078 KZC14498.1 KK853603 KDR06363.1 QOIP01000004 RLU23674.1 GBYB01012744 JAG82511.1 KZ288215 PBC32561.1 GEBQ01032004 JAT07973.1 GECU01024335 JAS83371.1 KK107046 EZA61723.1 KQ976424 KYM88478.1 ADTU01013057 GL763054 EFZ20207.1 GL887888 EGI69617.1 LBMM01001117 KMQ96893.1 DS233465 EDS30114.1
Proteomes
UP000218220
UP000007151
UP000183832
UP000075885
UP000007266
UP000008820
+ More
UP000075900 UP000095301 UP000000673 UP000075840 UP000075902 UP000075903 UP000268350 UP000037069 UP000076407 UP000007798 UP000008792 UP000076408 UP000069272 UP000002320 UP000000803 UP000007801 UP000008711 UP000001292 UP000001819 UP000000304 UP000091820 UP000009192 UP000092445 UP000078200 UP000192221 UP000002282 UP000001070 UP000095300 UP000008744 UP000192223 UP000092461 UP000092444 UP000007819 UP000053105 UP000076502 UP000027135 UP000279307 UP000242457 UP000079169 UP000053097 UP000078540 UP000005205 UP000007755 UP000036403
UP000075900 UP000095301 UP000000673 UP000075840 UP000075902 UP000075903 UP000268350 UP000037069 UP000076407 UP000007798 UP000008792 UP000076408 UP000069272 UP000002320 UP000000803 UP000007801 UP000008711 UP000001292 UP000001819 UP000000304 UP000091820 UP000009192 UP000092445 UP000078200 UP000192221 UP000002282 UP000001070 UP000095300 UP000008744 UP000192223 UP000092461 UP000092444 UP000007819 UP000053105 UP000076502 UP000027135 UP000279307 UP000242457 UP000079169 UP000053097 UP000078540 UP000005205 UP000007755 UP000036403
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K7E0
A0A212FHD2
U5EP92
A0A1J1J0M9
A0A336LL42
A0A182PL18
+ More
C0LTQ2 A0A1S4FNL5 Q16UK2 A0A182R7V4 T1PJK7 W5JW74 A0A182HMV0 A0A182UDD1 A0A182VPD7 A0A3B0J7J1 A0A0L0CQY4 A0A182XNL1 B4NQ77 A0A1Y1K121 B4M833 A0A2M4AG40 Q8SZ42 A0A182Y4V6 A0A182FMF6 B0XJN6 X2JBI9 A0A2M4AI58 B3MYZ1 B3NWS6 Q9VYG9 B4IGM6 A0A0K8UIM1 Q29H67 B4R418 A0A034VNU6 A0A1A9WAZ0 B4L6F9 A0A1B0AJ05 A0A1A9UHU7 A0A0A1WHB1 A0A1W4V7T5 B4Q211 B4JKL4 W8AUB2 A0A1I8NLM2 A0A1L8DPX4 B4H084 V5K5P8 Q2TJG0 V5K6B7 Q0ZS49 A0A1W4WFV5 C6FFT9 A0A1B0CWT6 A0A1B0FM16 J9JV77 A0A2H8TEH8 A0A2S2N722 A0A1B6DBG7 A0A0N0BF21 A0A154PT88 A0A067QSN2 A0A3L8DTG3 A0A0C9R8Q7 A0A2A3ENE0 A0A1S3D1V7 A0A1B6K949 A0A1B6I8W4 A0A026X0W9 A0A195BQR4 A0A158NDW9 E9IGS3 F4W7U7 A0A0J7L2K4 B0XJ99
C0LTQ2 A0A1S4FNL5 Q16UK2 A0A182R7V4 T1PJK7 W5JW74 A0A182HMV0 A0A182UDD1 A0A182VPD7 A0A3B0J7J1 A0A0L0CQY4 A0A182XNL1 B4NQ77 A0A1Y1K121 B4M833 A0A2M4AG40 Q8SZ42 A0A182Y4V6 A0A182FMF6 B0XJN6 X2JBI9 A0A2M4AI58 B3MYZ1 B3NWS6 Q9VYG9 B4IGM6 A0A0K8UIM1 Q29H67 B4R418 A0A034VNU6 A0A1A9WAZ0 B4L6F9 A0A1B0AJ05 A0A1A9UHU7 A0A0A1WHB1 A0A1W4V7T5 B4Q211 B4JKL4 W8AUB2 A0A1I8NLM2 A0A1L8DPX4 B4H084 V5K5P8 Q2TJG0 V5K6B7 Q0ZS49 A0A1W4WFV5 C6FFT9 A0A1B0CWT6 A0A1B0FM16 J9JV77 A0A2H8TEH8 A0A2S2N722 A0A1B6DBG7 A0A0N0BF21 A0A154PT88 A0A067QSN2 A0A3L8DTG3 A0A0C9R8Q7 A0A2A3ENE0 A0A1S3D1V7 A0A1B6K949 A0A1B6I8W4 A0A026X0W9 A0A195BQR4 A0A158NDW9 E9IGS3 F4W7U7 A0A0J7L2K4 B0XJ99
PDB
1POC
E-value=2.79183e-18,
Score=225
Ontologies
GO
Topology
Subcellular location
Secreted
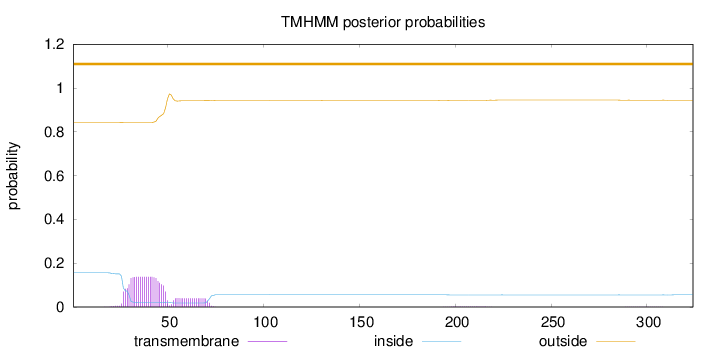
Length:
324
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.64801000000001
Exp number, first 60 AAs:
3.15483
Total prob of N-in:
0.15661
outside
1 - 324
Population Genetic Test Statistics
Pi
20.457791
Theta
17.615404
Tajima's D
-1.967779
CLR
1.702829
CSRT
0.0164991750412479
Interpretation
Uncertain
