Gene
KWMTBOMO04808
Pre Gene Modal
BGIBMGA009912
Annotation
PREDICTED:_protein-S-isoprenylcysteine_O-methyltransferase_[Papilio_machaon]
Full name
Protein-S-isoprenylcysteine O-methyltransferase
Location in the cell
PlasmaMembrane Reliability : 4.966
Sequence
CDS
ATGAAAAACATATGCCCCGCCGGAAAACAAGCGATAATATGGTTTATGCTAACCGTTTGTATATTTTTCATATCATTCTTTTCTGGAAGCCTTTTAGGATTCACATCTGAGATCTGGGCACTTACTTATTGGGGTCCCTCATTATATTTCTGTTTGTTGAATTTTATTTTAAGATATGCCTATAAAGGATTTTTATATGAGGTGTCAATACGAGCAGCATTCCTCGGAGCAGTATTCACAATAGGCCTCTATTTATCGACATTTGAAGATGGCATTAAGGTTTTCGGTCTCTACACCATGGTGCTGTCGATGTTCCATTTTTCTGAGTTCATGAGTGTAGCCCTAACAAACCCTAGGACATTAACAATTGATTCATATATATTGAATCACAGTGTGCAGTATGGAGTTGCAGCGGTCACTAGTTGGATTGAATGGGCCATTGAATATTATTTCTTTCCGGGGATGAAGACTTATTTCTGGTTGTCAAGCATAGGTGTAATAATGTGTATAGGAGGAGAAATATTAAGAAAATCAGCTATGTTCACAGCCAAAACTAACTTCAACCACACAGTACAATTCGTAAAGAGAGTGGATCATCGGCTAGTTACGCACGGGGTGTACTCTCTCTGTAGACATCCAAGCTATGTAGGATGGTTCTATTGGTCAGTCGGTACTCAGTTTATCCTCCTGAATCCTTTTTGCCTGATCATCTACACGCTAGCCTCGTGGATGTTCTTTAGGGAAAGAGTTTATGCTGAGGAATTGACACTATTGACATTCTTTGGTCCCGCCTATGAAGTCTACCAAAGCAAAGTCAGTACAGGCCTTCCGTTCATCCAGGGTTATGTACCAGAGAACACAGGACAATTTGTCAAAGAAGACCATTGGAGTTACTGA
Protein
MKNICPAGKQAIIWFMLTVCIFFISFFSGSLLGFTSEIWALTYWGPSLYFCLLNFILRYAYKGFLYEVSIRAAFLGAVFTIGLYLSTFEDGIKVFGLYTMVLSMFHFSEFMSVALTNPRTLTIDSYILNHSVQYGVAAVTSWIEWAIEYYFFPGMKTYFWLSSIGVIMCIGGEILRKSAMFTAKTNFNHTVQFVKRVDHRLVTHGVYSLCRHPSYVGWFYWSVGTQFILLNPFCLIIYTLASWMFFRERVYAEELTLLTFFGPAYEVYQSKVSTGLPFIQGYVPENTGQFVKEDHWSY
Summary
Catalytic Activity
[protein]-C-terminal S-[(2E,6E)-farnesyl]-L-cysteine + S-adenosyl-L-methionine = [protein]-C-terminal S-[(2E,6E)-farnesyl]-L-cysteine methyl ester + S-adenosyl-L-homocysteine
Cofactor
Zn(2+)
Similarity
Belongs to the class VI-like SAM-binding methyltransferase superfamily. Isoprenylcysteine carboxyl methyltransferase family.
Uniprot
H9JK61
A0A2H1WV57
A0A3S2LWB9
A0A194RCV9
A0A194PR43
A0A2A4K7M7
+ More
A0A212FHF2 A0A1E1W8E9 A0A1W4WHL0 A0A067R9D3 A0A0L0BUD4 A0A0L7QSP4 A0A1B6EU79 A0A1B6EVX0 A0A026X1E4 A0A1B6J5S1 A0A1I8PFP1 B3NCY3 B4L0J4 A0A1B6KK98 A0A1I8MQ03 B4PHB2 B4HGN4 A0A1L8DTQ6 E9IR42 B3MAU2 A0A3B0JS31 B4QIY4 A0A069DRE1 A0A224XJ06 Q2M0C1 B4GRU3 A0A1W4VJJ0 A0A195E430 Q9VU37 A0A0A1XI35 A0A154PRU5 A0A2A3EED2 A0A0K8WG38 E2AWE2 A0A195C132 F4WJM6 A0A0V0G8H3 B4LBH3 A0A3R7MGL4 A0A195FWB1 K1PPT4 A0A195AUC0 U5EKS2 A0A158NNC1 A0A1J1HXB0 A0A1A9ZKP5 D3TNL2 B4MM66 A0A1A9UFT9 E2BPW0 A0A1A9XGE7 A0A1B3PDH5 A0A232EXP3 A0A2P2I3B3 A0A1B0AWF9 B4K006 A0A182JF20 A0A0P4WHV6 A0A182FRK2 A0A210PXN4 A0A310S4W4 V3ZGY2 A0A023F474 A0A182YBD6 A0A1S4FUR4 A0A182P9K3 A0A084VXV7 A0A1Q3F6H3 A0A0K8TSQ5 T1HDA1 A0A131XYF9 W5J3Q6 A0A336KG15 Q16NH8 U4TVP3 U3FCE4 A0A2M4AXT5 A0A182UMX8 A0A182X9L2 A0A182L8T7 A0A182HKJ5 Q7PXA7 A0A0B8RX18 A0A0M4EER0 A0A1S3JQC1 A0A182Q4P4 A0A1A9WSN9 A0A182NQK0 A0A0L8HV91 A0A098LXW4 E0VRB9 Q6INK2 A0A182GTA5
A0A212FHF2 A0A1E1W8E9 A0A1W4WHL0 A0A067R9D3 A0A0L0BUD4 A0A0L7QSP4 A0A1B6EU79 A0A1B6EVX0 A0A026X1E4 A0A1B6J5S1 A0A1I8PFP1 B3NCY3 B4L0J4 A0A1B6KK98 A0A1I8MQ03 B4PHB2 B4HGN4 A0A1L8DTQ6 E9IR42 B3MAU2 A0A3B0JS31 B4QIY4 A0A069DRE1 A0A224XJ06 Q2M0C1 B4GRU3 A0A1W4VJJ0 A0A195E430 Q9VU37 A0A0A1XI35 A0A154PRU5 A0A2A3EED2 A0A0K8WG38 E2AWE2 A0A195C132 F4WJM6 A0A0V0G8H3 B4LBH3 A0A3R7MGL4 A0A195FWB1 K1PPT4 A0A195AUC0 U5EKS2 A0A158NNC1 A0A1J1HXB0 A0A1A9ZKP5 D3TNL2 B4MM66 A0A1A9UFT9 E2BPW0 A0A1A9XGE7 A0A1B3PDH5 A0A232EXP3 A0A2P2I3B3 A0A1B0AWF9 B4K006 A0A182JF20 A0A0P4WHV6 A0A182FRK2 A0A210PXN4 A0A310S4W4 V3ZGY2 A0A023F474 A0A182YBD6 A0A1S4FUR4 A0A182P9K3 A0A084VXV7 A0A1Q3F6H3 A0A0K8TSQ5 T1HDA1 A0A131XYF9 W5J3Q6 A0A336KG15 Q16NH8 U4TVP3 U3FCE4 A0A2M4AXT5 A0A182UMX8 A0A182X9L2 A0A182L8T7 A0A182HKJ5 Q7PXA7 A0A0B8RX18 A0A0M4EER0 A0A1S3JQC1 A0A182Q4P4 A0A1A9WSN9 A0A182NQK0 A0A0L8HV91 A0A098LXW4 E0VRB9 Q6INK2 A0A182GTA5
EC Number
2.1.1.100
Pubmed
19121390
26354079
22118469
24845553
26108605
24508170
+ More
30249741 17994087 25315136 17550304 21282665 22936249 26334808 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 20798317 21719571 22992520 21347285 20353571 28648823 28812685 23254933 25474469 25244985 24438588 26369729 29652888 20920257 23761445 17510324 23537049 23915248 20966253 12364791 14747013 17210077 25476704 20566863 26483478
30249741 17994087 25315136 17550304 21282665 22936249 26334808 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 20798317 21719571 22992520 21347285 20353571 28648823 28812685 23254933 25474469 25244985 24438588 26369729 29652888 20920257 23761445 17510324 23537049 23915248 20966253 12364791 14747013 17210077 25476704 20566863 26483478
EMBL
BABH01026710
BABH01026711
BABH01026712
ODYU01011250
SOQ56872.1
RSAL01000151
+ More
RVE45756.1 KQ460367 KPJ15457.1 KQ459595 KPI95782.1 NWSH01000074 PCG79938.1 AGBW02008504 OWR53154.1 GDQN01007788 JAT83266.1 KK852804 KDR16214.1 JRES01001310 KNC23685.1 KQ414756 KOC61662.1 GECZ01028224 JAS41545.1 GECZ01027644 JAS42125.1 KK107031 QOIP01000008 EZA62100.1 RLU19318.1 GECU01013199 JAS94507.1 CH954178 EDV51639.1 CH933809 EDW19163.1 GEBQ01028111 JAT11866.1 CM000159 EDW94373.2 CH480815 EDW41342.1 GFDF01004266 JAV09818.1 GL765043 EFZ16968.1 CH902618 EDV39176.1 OUUW01000002 SPP76166.1 CM000363 CM002912 EDX10315.1 KMY99349.1 GBGD01002429 JAC86460.1 GFTR01003950 JAW12476.1 CH379069 EAL31013.1 CH479188 EDW40478.1 KQ979685 KYN19841.1 AE014296 AY070700 AY071003 AAF49854.1 AAL48171.1 AGB94486.1 GBXI01003651 JAD10641.1 KQ435090 KZC14593.1 KZ288266 PBC30143.1 GDHF01002509 JAI49805.1 GL443286 EFN62242.1 KQ978457 KYM93878.1 GL888186 EGI65518.1 GECL01001665 JAP04459.1 CH940647 EDW70783.1 QCYY01001711 ROT75935.1 KQ981208 KYN44711.1 JH818211 EKC18440.1 KQ976738 KYM75833.1 GANO01001698 JAB58173.1 ADTU01021232 CVRI01000021 CRK91988.1 CCAG010012835 EZ423014 ADD19290.1 CH963847 EDW73075.1 GL449658 EFN82311.1 KU659973 AOG17771.1 NNAY01001723 OXU23123.1 IACF01002889 LAB68524.1 JXJN01004688 CH916383 EDV98688.1 GDRN01050903 JAI66435.1 NEDP02005415 OWF41250.1 KQ773953 OAD52337.1 KB202408 ESO90488.1 GBBI01002903 JAC15809.1 ATLV01018208 KE525224 KFB42801.1 GFDL01011898 JAV23147.1 GDAI01000447 JAI17156.1 ACPB03001054 GEFM01004531 GEGO01002690 JAP71265.1 JAR92714.1 ADMH02002105 ETN59002.1 UFQS01000320 UFQT01000320 SSX02852.1 SSX23220.1 CH477823 EAT35916.1 KB631628 ERL84837.1 GAEP01000772 JAB54049.1 GGFK01012295 MBW45616.1 APCN01000834 AAAB01008987 EAA01195.3 GBSH01002548 JAG66478.1 CP012525 ALC44308.1 AXCN02001518 KQ417224 KOF93138.1 GBSI01001193 JAC95303.1 DS235465 EEB15925.1 BC072278 AAH72278.1 JXUM01086651 JXUM01086652 KQ563582 KXJ73635.1
RVE45756.1 KQ460367 KPJ15457.1 KQ459595 KPI95782.1 NWSH01000074 PCG79938.1 AGBW02008504 OWR53154.1 GDQN01007788 JAT83266.1 KK852804 KDR16214.1 JRES01001310 KNC23685.1 KQ414756 KOC61662.1 GECZ01028224 JAS41545.1 GECZ01027644 JAS42125.1 KK107031 QOIP01000008 EZA62100.1 RLU19318.1 GECU01013199 JAS94507.1 CH954178 EDV51639.1 CH933809 EDW19163.1 GEBQ01028111 JAT11866.1 CM000159 EDW94373.2 CH480815 EDW41342.1 GFDF01004266 JAV09818.1 GL765043 EFZ16968.1 CH902618 EDV39176.1 OUUW01000002 SPP76166.1 CM000363 CM002912 EDX10315.1 KMY99349.1 GBGD01002429 JAC86460.1 GFTR01003950 JAW12476.1 CH379069 EAL31013.1 CH479188 EDW40478.1 KQ979685 KYN19841.1 AE014296 AY070700 AY071003 AAF49854.1 AAL48171.1 AGB94486.1 GBXI01003651 JAD10641.1 KQ435090 KZC14593.1 KZ288266 PBC30143.1 GDHF01002509 JAI49805.1 GL443286 EFN62242.1 KQ978457 KYM93878.1 GL888186 EGI65518.1 GECL01001665 JAP04459.1 CH940647 EDW70783.1 QCYY01001711 ROT75935.1 KQ981208 KYN44711.1 JH818211 EKC18440.1 KQ976738 KYM75833.1 GANO01001698 JAB58173.1 ADTU01021232 CVRI01000021 CRK91988.1 CCAG010012835 EZ423014 ADD19290.1 CH963847 EDW73075.1 GL449658 EFN82311.1 KU659973 AOG17771.1 NNAY01001723 OXU23123.1 IACF01002889 LAB68524.1 JXJN01004688 CH916383 EDV98688.1 GDRN01050903 JAI66435.1 NEDP02005415 OWF41250.1 KQ773953 OAD52337.1 KB202408 ESO90488.1 GBBI01002903 JAC15809.1 ATLV01018208 KE525224 KFB42801.1 GFDL01011898 JAV23147.1 GDAI01000447 JAI17156.1 ACPB03001054 GEFM01004531 GEGO01002690 JAP71265.1 JAR92714.1 ADMH02002105 ETN59002.1 UFQS01000320 UFQT01000320 SSX02852.1 SSX23220.1 CH477823 EAT35916.1 KB631628 ERL84837.1 GAEP01000772 JAB54049.1 GGFK01012295 MBW45616.1 APCN01000834 AAAB01008987 EAA01195.3 GBSH01002548 JAG66478.1 CP012525 ALC44308.1 AXCN02001518 KQ417224 KOF93138.1 GBSI01001193 JAC95303.1 DS235465 EEB15925.1 BC072278 AAH72278.1 JXUM01086651 JXUM01086652 KQ563582 KXJ73635.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000218220
UP000007151
+ More
UP000192223 UP000027135 UP000037069 UP000053825 UP000053097 UP000279307 UP000095300 UP000008711 UP000009192 UP000095301 UP000002282 UP000001292 UP000007801 UP000268350 UP000000304 UP000001819 UP000008744 UP000192221 UP000078492 UP000000803 UP000076502 UP000242457 UP000000311 UP000078542 UP000007755 UP000008792 UP000283509 UP000078541 UP000005408 UP000078540 UP000005205 UP000183832 UP000092445 UP000092444 UP000007798 UP000078200 UP000008237 UP000092443 UP000215335 UP000092460 UP000001070 UP000075880 UP000069272 UP000242188 UP000030746 UP000076408 UP000075885 UP000030765 UP000015103 UP000000673 UP000008820 UP000030742 UP000075903 UP000076407 UP000075882 UP000075840 UP000007062 UP000092553 UP000085678 UP000075886 UP000091820 UP000075884 UP000053454 UP000009046 UP000069940 UP000249989
UP000192223 UP000027135 UP000037069 UP000053825 UP000053097 UP000279307 UP000095300 UP000008711 UP000009192 UP000095301 UP000002282 UP000001292 UP000007801 UP000268350 UP000000304 UP000001819 UP000008744 UP000192221 UP000078492 UP000000803 UP000076502 UP000242457 UP000000311 UP000078542 UP000007755 UP000008792 UP000283509 UP000078541 UP000005408 UP000078540 UP000005205 UP000183832 UP000092445 UP000092444 UP000007798 UP000078200 UP000008237 UP000092443 UP000215335 UP000092460 UP000001070 UP000075880 UP000069272 UP000242188 UP000030746 UP000076408 UP000075885 UP000030765 UP000015103 UP000000673 UP000008820 UP000030742 UP000075903 UP000076407 UP000075882 UP000075840 UP000007062 UP000092553 UP000085678 UP000075886 UP000091820 UP000075884 UP000053454 UP000009046 UP000069940 UP000249989
Pfam
PF04140 ICMT
ProteinModelPortal
H9JK61
A0A2H1WV57
A0A3S2LWB9
A0A194RCV9
A0A194PR43
A0A2A4K7M7
+ More
A0A212FHF2 A0A1E1W8E9 A0A1W4WHL0 A0A067R9D3 A0A0L0BUD4 A0A0L7QSP4 A0A1B6EU79 A0A1B6EVX0 A0A026X1E4 A0A1B6J5S1 A0A1I8PFP1 B3NCY3 B4L0J4 A0A1B6KK98 A0A1I8MQ03 B4PHB2 B4HGN4 A0A1L8DTQ6 E9IR42 B3MAU2 A0A3B0JS31 B4QIY4 A0A069DRE1 A0A224XJ06 Q2M0C1 B4GRU3 A0A1W4VJJ0 A0A195E430 Q9VU37 A0A0A1XI35 A0A154PRU5 A0A2A3EED2 A0A0K8WG38 E2AWE2 A0A195C132 F4WJM6 A0A0V0G8H3 B4LBH3 A0A3R7MGL4 A0A195FWB1 K1PPT4 A0A195AUC0 U5EKS2 A0A158NNC1 A0A1J1HXB0 A0A1A9ZKP5 D3TNL2 B4MM66 A0A1A9UFT9 E2BPW0 A0A1A9XGE7 A0A1B3PDH5 A0A232EXP3 A0A2P2I3B3 A0A1B0AWF9 B4K006 A0A182JF20 A0A0P4WHV6 A0A182FRK2 A0A210PXN4 A0A310S4W4 V3ZGY2 A0A023F474 A0A182YBD6 A0A1S4FUR4 A0A182P9K3 A0A084VXV7 A0A1Q3F6H3 A0A0K8TSQ5 T1HDA1 A0A131XYF9 W5J3Q6 A0A336KG15 Q16NH8 U4TVP3 U3FCE4 A0A2M4AXT5 A0A182UMX8 A0A182X9L2 A0A182L8T7 A0A182HKJ5 Q7PXA7 A0A0B8RX18 A0A0M4EER0 A0A1S3JQC1 A0A182Q4P4 A0A1A9WSN9 A0A182NQK0 A0A0L8HV91 A0A098LXW4 E0VRB9 Q6INK2 A0A182GTA5
A0A212FHF2 A0A1E1W8E9 A0A1W4WHL0 A0A067R9D3 A0A0L0BUD4 A0A0L7QSP4 A0A1B6EU79 A0A1B6EVX0 A0A026X1E4 A0A1B6J5S1 A0A1I8PFP1 B3NCY3 B4L0J4 A0A1B6KK98 A0A1I8MQ03 B4PHB2 B4HGN4 A0A1L8DTQ6 E9IR42 B3MAU2 A0A3B0JS31 B4QIY4 A0A069DRE1 A0A224XJ06 Q2M0C1 B4GRU3 A0A1W4VJJ0 A0A195E430 Q9VU37 A0A0A1XI35 A0A154PRU5 A0A2A3EED2 A0A0K8WG38 E2AWE2 A0A195C132 F4WJM6 A0A0V0G8H3 B4LBH3 A0A3R7MGL4 A0A195FWB1 K1PPT4 A0A195AUC0 U5EKS2 A0A158NNC1 A0A1J1HXB0 A0A1A9ZKP5 D3TNL2 B4MM66 A0A1A9UFT9 E2BPW0 A0A1A9XGE7 A0A1B3PDH5 A0A232EXP3 A0A2P2I3B3 A0A1B0AWF9 B4K006 A0A182JF20 A0A0P4WHV6 A0A182FRK2 A0A210PXN4 A0A310S4W4 V3ZGY2 A0A023F474 A0A182YBD6 A0A1S4FUR4 A0A182P9K3 A0A084VXV7 A0A1Q3F6H3 A0A0K8TSQ5 T1HDA1 A0A131XYF9 W5J3Q6 A0A336KG15 Q16NH8 U4TVP3 U3FCE4 A0A2M4AXT5 A0A182UMX8 A0A182X9L2 A0A182L8T7 A0A182HKJ5 Q7PXA7 A0A0B8RX18 A0A0M4EER0 A0A1S3JQC1 A0A182Q4P4 A0A1A9WSN9 A0A182NQK0 A0A0L8HV91 A0A098LXW4 E0VRB9 Q6INK2 A0A182GTA5
PDB
5V7P
E-value=9.36871e-62,
Score=599
Ontologies
GO
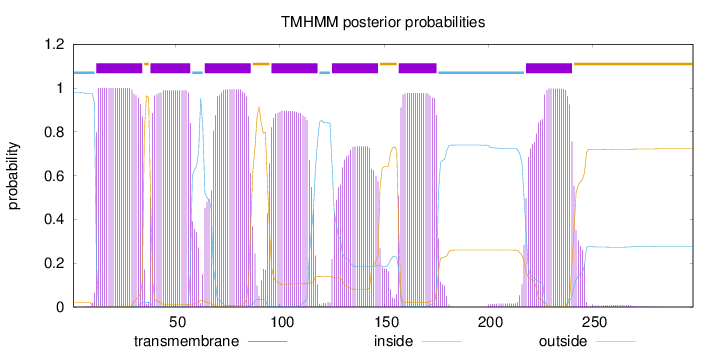
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
298
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
140.9543
Exp number, first 60 AAs:
42.83703
Total prob of N-in:
0.97829
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 37
TMhelix
38 - 57
inside
58 - 63
TMhelix
64 - 86
outside
87 - 95
TMhelix
96 - 118
inside
119 - 124
TMhelix
125 - 147
outside
148 - 156
TMhelix
157 - 175
inside
176 - 217
TMhelix
218 - 240
outside
241 - 298
Population Genetic Test Statistics
Pi
26.153712
Theta
25.531187
Tajima's D
0.134316
CLR
0.557037
CSRT
0.410729463526824
Interpretation
Uncertain
