Gene
KWMTBOMO04806 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009911
Annotation
PREDICTED:_ubiquitin-like_modifier-activating_enzyme_1_isoform_X1_[Papilio_machaon]
Full name
DNA ligase
Location in the cell
Cytoplasmic Reliability : 2.975
Sequence
CDS
ATGTCTAGTGCTGAAGTCGCCGATAATTCCGTTGACCCCCCGGCGAAAAAGCGGAAACTAAATACAGGAGAAGCGCATTCGAACAACTCGGCAATGGCGAACAATGGAGCGCGCGTGGAAGATGAAATCGACGAGAGTCTGTACTCGCGCCAGCTCTACGTGCTGGGCCACGACGCTATGCGTCGAATGGCGAGTTCTGATGTCCTTATATCTGGCCTTGGCGGTTTAGGTGTCGAAATAGCGAAAAACGTTATCCTTGGTGGTGTAAAGTCGGTAACTTTGCATGACGACAAAAATTGCACCGTGGCTGATCTTTCCTCGCAGTTCTACCTTTCAGAGACTGTAATTGGTCAGAACAGGGCTTTAACATCGTGCGAACAACTTTCTGAACTGAACCACTATGTGCCCACTACTGCTTACACAGGACCTTTAACTGAAGACTTTTTGCGTAAGTTCCGTGTTGTGGTCCTCACAGGAGCATCGTGGGCTGAGCAGGAGCGAGTAGCTGCGTTCACTCACGCCAATAACATAGCATTGGTCATTGCTGATACTAGAGGCCTGTTCTCACAAGTGTTCTGTGACTTCGGACCAGAGTTTACGGTGTTAGATGTAAATGGAGAGAATCCAGTCTCAGCTATGATTGCAGCCATTACGCAAGACTATGAGGCAGTTGTGACATGTTTGGATGATACTCGTCACGGATTGGAGGATGGCGACTATGTTACGTTTAGTGAGGTTCAAGGTATGACAGAACTAAATGGCTGTGAACCCCGCAAGATCAAAGTCCTTGGTCCCTACACTTTTAGTATAGGTGATACTACAAACTTCTCCAAATACATCCGAGGAGGTATTGTTACACAGGTCAAAATGCCCAAGAAGATCAACTTCAAGCCTTTGAGTGAATCATTCAAACAACCAGAATTCTTGATCTCTGATTTTGCCAAGTTTGAATATCCACAGCAGTTGCATTTGGCTTTTGCTGCTCTTCACAAGTTCCAGGCTGGAGAAGGTCGTCTGCCAAAGCCATGGAATGATGAAGATGTGGCAAAATTCAAGCTGTATGTGAAAACTGTTCTAGAGGATGGACTTTATAAGGATGAAGATTTTGAAGTGAATACTGAATTATTGGAAACATTCTGTAAGATATCGTCAGGAGACCTAAATCCTATGAACGCAGCGATAGGCGGTGTGGTTGCTCAAGAGGTAATGAAGGCTTGTTCTGGCAAGTTCCACCCACTTGTACAGTGGCTTTACTTGGATGCTATTGAGTGCTTACCTAAAGACCATTCAACTCTCACCGAAGAAAATTGCAAACCAGTCGGTTCACGATATGATGGACAGGTAGCTGTTTTTGGTAGAGAATTCCAAAAGAAACTTGGACAGCTGAAGTATTTTATTGTGGGTGCTGGTGCTATCGGATGTGAATTATTAAAGAACTTTGCCATGATTGGTGTGGGTGCTGATGGAGGACAAATTACAGTCACGGATATGGACCTCATTGAGAAGTCTAACCTCAACAGGCAGTTCTTGTTTAGGCCTTATGATGTACAGAAACCGAAATCTAGTACTGCAGCTAATGTAATTAAACGCATGAATCCAGCAGTAAATGTTTTGGCACAAGAAAACAGAGTTTGCCCTGAGACCGAGTCCGTCTACAATGATGCATTCTTTGAAGCATTGGATGGTGTAGCCAATGCACTCGACAATGTCGACGCCCGTATATACATGGACAGACGGTGTGTGTATTATAGAAAACCTTTATTGGAGAGTGGCACTCTTGGAACTAAAGGAAATACACAGGTGGTTGTTCCATTCCTTACGGAATCTTACAGCTCATCCCAGGACCCTCCCGAGAAGAGTATTCCTATTTGTACGCTCAAGAACTTCCCGAATGCCATTGAACACACACTACAATGGGCTCGCGATGAATTCGAAGGTTTATTCCGTCAGGCTGCTGAGCATGCTGCCCAATACTTGCAGGACCCGAATTTCTTGGACCGCTGTCTTAAATTGCCTGGTACTCAGCCTTTGGATGCTATTGAGAGTGTGCGAAACGCTATAAACGAACGCCCTCACAGTTTCGACGACTGCGTGATATGGGCGAGACATCATTGGGAGAACCAATACGCGAACCAGATCAAGCAGCTCCTTTACAACTTCCCGGCGAAACAGTTGACCACGAGCGGAGCGCCTTTCTGGTCCGGACCGAAGCGTTGTCCTTCACCCCTCGCCTTCGATCCTAATGATGAACTACACGTCGATTACATTGTTGCCGCAGCGAACCTCAGGGCTACGGTATATGGAATCCCATCCTGTGTTGACAGGGAAAACATCGCCAAGCTTGCCGCTAGCATACAGGTACCGAAATTCAGCCCCAAATCCGGCGTGAAGATCGCGGTGAGCGACGCCCAGTTACAGCAAAACAATGACGAGATGGATCAAGACAGGGTTAAGAACATGATCGCAGAGCTGCCGTCTCCAAGCAAACTCGGATCTCTTAAGATCACACCGCTAGAATTCGAAAAAGATGATGATACAAATTTCCACATGGACTTCATCGTTGCCGCCTCCAACCTGCGGGCTACCAACTACAAGATCACGCCAGCCGACAGACATCGCTCTAAACTCATCGCTGGCAAGATTATCCCTGCCATTGCCACTACAACATCCGTGGTGGCTGGCTTAGTCTGCCTCGAACTTTACAAACTGGCCCAAGGCTTCAATAAGCTGGAAGTGTTCAAGAACGGCTTCGTCAACCTTGCCCTGCCCTTCTTCGGTTTCTCTGAACCGATTGCGCCGTCGACCAACACCTACAATGAGAATAAGTGGACATTATGGGACAGATTTGAAGTTAAAGGAGAGATTACATTACAGCAATTTTTAGATCACTTCAAAAACGAACACAAGTTGGAGATCACCATGTTGTCTCAAGGTGTGTGCATGCTTTATTCGTTCTTCATGCCCAAAGCCAAGCGGCTGGAACGATTGAACCTGCCAATGTCGGAGGTGGTCACAAAAGTGTCCAAGAAGAAGTTGGAACCGCATGTGAATGCATTAGTTTTTGAATTGTGCTGCAATGATGATGACGGGAATGATGTGGAGGTCCCGTATGTAAAATACACTCTGCCTTAG
Protein
MSSAEVADNSVDPPAKKRKLNTGEAHSNNSAMANNGARVEDEIDESLYSRQLYVLGHDAMRRMASSDVLISGLGGLGVEIAKNVILGGVKSVTLHDDKNCTVADLSSQFYLSETVIGQNRALTSCEQLSELNHYVPTTAYTGPLTEDFLRKFRVVVLTGASWAEQERVAAFTHANNIALVIADTRGLFSQVFCDFGPEFTVLDVNGENPVSAMIAAITQDYEAVVTCLDDTRHGLEDGDYVTFSEVQGMTELNGCEPRKIKVLGPYTFSIGDTTNFSKYIRGGIVTQVKMPKKINFKPLSESFKQPEFLISDFAKFEYPQQLHLAFAALHKFQAGEGRLPKPWNDEDVAKFKLYVKTVLEDGLYKDEDFEVNTELLETFCKISSGDLNPMNAAIGGVVAQEVMKACSGKFHPLVQWLYLDAIECLPKDHSTLTEENCKPVGSRYDGQVAVFGREFQKKLGQLKYFIVGAGAIGCELLKNFAMIGVGADGGQITVTDMDLIEKSNLNRQFLFRPYDVQKPKSSTAANVIKRMNPAVNVLAQENRVCPETESVYNDAFFEALDGVANALDNVDARIYMDRRCVYYRKPLLESGTLGTKGNTQVVVPFLTESYSSSQDPPEKSIPICTLKNFPNAIEHTLQWARDEFEGLFRQAAEHAAQYLQDPNFLDRCLKLPGTQPLDAIESVRNAINERPHSFDDCVIWARHHWENQYANQIKQLLYNFPAKQLTTSGAPFWSGPKRCPSPLAFDPNDELHVDYIVAAANLRATVYGIPSCVDRENIAKLAASIQVPKFSPKSGVKIAVSDAQLQQNNDEMDQDRVKNMIAELPSPSKLGSLKITPLEFEKDDDTNFHMDFIVAASNLRATNYKITPADRHRSKLIAGKIIPAIATTTSVVAGLVCLELYKLAQGFNKLEVFKNGFVNLALPFFGFSEPIAPSTNTYNENKWTLWDRFEVKGEITLQQFLDHFKNEHKLEITMLSQGVCMLYSFFMPKAKRLERLNLPMSEVVTKVSKKKLEPHVNALVFELCCNDDDGNDVEVPYVKYTLP
Summary
Catalytic Activity
ATP + (deoxyribonucleotide)(n)-3'-hydroxyl + 5'-phospho-(deoxyribonucleotide)(m) = (deoxyribonucleotide)(n+m) + AMP + diphosphate.
Similarity
Belongs to the ubiquitin-activating E1 family.
Belongs to the ATP-dependent DNA ligase family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the ATP-dependent DNA ligase family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
H9JK60
A0A2H1X2R9
A0A2A4K8C9
A0A3S2PA79
A0A194PXA9
S4PKG8
+ More
A0A212FHF4 A0A139WLH6 A0A1B6EJ56 A0A1B6J6R3 A0A2J7QBF8 A0A2P8XPS9 A0A2R7VVH7 A0A067RA93 A0A2D1QUE3 A0A0M8ZPK0 A0A158NEC8 A0A195BW67 R4WDT6 A0A026W156 A0A151J6R2 F4WJ85 A0A195FM65 A0A1W4WLZ4 A0A194RDB5 A0A1W4WBV3 A0A088AVS9 A0A310SJ05 A0A2A3EPK1 V9IJG1 A0A195CCB1 A0A154PPG8 A0A151WVW5 E2C890 A0A0L7RIX0 A0A0A9XZ86 A0A1Y1K0A8 A0A0K8SD12 U4UXQ1 A0A0V0G561 A0A0P4VU82 A0A0J7P381 A0A069DXS0 A0A0C9RUB8 A0A0T6AZP6 A0A224XIR4 T1HIR2 N6TVS5 U5EX66 A0A1B6D3R9 A0A1L8DTD2 A0A1Q3F816 A0A1Q3F861 A0A1Q3F804 A0A1Q3F854 A0A1Q3F820 A0A2S2NJW1 B0W2P8 A0A023EWP6 A0A0P5RDB7 A0A164KX14 A0A2H8TID4 A0A182QD44 A0A1B2JLV6 J9JZV9 Q17N85 A0A0P4WEI8 A0A182JRA0 A0A2M4BBE8 A0A2M4BC15 A0A2C9GQF2 A0A2S2Q4C1 Q17N86 M9NPG8 A0A2M3ZFB7 A0A182XJW3 A0A182UP22 A0A2M3YYE4 A0A182Y9D9 A0A2M3YYE8 E9FRU9 A0A131Z2F0 A0A1S4H7Q4 L7M9E9 A0A0P4WA61 A0A182PKF3 A0A1E1X320 A0A182JD16 A0A2M4A016 W5J6E3 A0A2M3ZZU6 A0A2M4CQJ6 A0A182RRY5 A0A023G0D6 A0A386H7J3 G3MM20 A0A1Y9H2R6 A0A131XSJ1 A0A131XEH3 A0A182FBD8
A0A212FHF4 A0A139WLH6 A0A1B6EJ56 A0A1B6J6R3 A0A2J7QBF8 A0A2P8XPS9 A0A2R7VVH7 A0A067RA93 A0A2D1QUE3 A0A0M8ZPK0 A0A158NEC8 A0A195BW67 R4WDT6 A0A026W156 A0A151J6R2 F4WJ85 A0A195FM65 A0A1W4WLZ4 A0A194RDB5 A0A1W4WBV3 A0A088AVS9 A0A310SJ05 A0A2A3EPK1 V9IJG1 A0A195CCB1 A0A154PPG8 A0A151WVW5 E2C890 A0A0L7RIX0 A0A0A9XZ86 A0A1Y1K0A8 A0A0K8SD12 U4UXQ1 A0A0V0G561 A0A0P4VU82 A0A0J7P381 A0A069DXS0 A0A0C9RUB8 A0A0T6AZP6 A0A224XIR4 T1HIR2 N6TVS5 U5EX66 A0A1B6D3R9 A0A1L8DTD2 A0A1Q3F816 A0A1Q3F861 A0A1Q3F804 A0A1Q3F854 A0A1Q3F820 A0A2S2NJW1 B0W2P8 A0A023EWP6 A0A0P5RDB7 A0A164KX14 A0A2H8TID4 A0A182QD44 A0A1B2JLV6 J9JZV9 Q17N85 A0A0P4WEI8 A0A182JRA0 A0A2M4BBE8 A0A2M4BC15 A0A2C9GQF2 A0A2S2Q4C1 Q17N86 M9NPG8 A0A2M3ZFB7 A0A182XJW3 A0A182UP22 A0A2M3YYE4 A0A182Y9D9 A0A2M3YYE8 E9FRU9 A0A131Z2F0 A0A1S4H7Q4 L7M9E9 A0A0P4WA61 A0A182PKF3 A0A1E1X320 A0A182JD16 A0A2M4A016 W5J6E3 A0A2M3ZZU6 A0A2M4CQJ6 A0A182RRY5 A0A023G0D6 A0A386H7J3 G3MM20 A0A1Y9H2R6 A0A131XSJ1 A0A131XEH3 A0A182FBD8
EC Number
6.5.1.1
Pubmed
EMBL
BABH01026707
ODYU01012959
SOQ59516.1
NWSH01000074
PCG79922.1
RSAL01000151
+ More
RVE45758.1 KQ459595 KPI95780.1 GAIX01004505 JAA88055.1 AGBW02008504 OWR53156.1 KQ971319 KYB28908.1 GECZ01031811 GECZ01030264 GECZ01026537 GECZ01022093 GECZ01017859 GECZ01014533 GECZ01012094 GECZ01001736 GECZ01001077 JAS37958.1 JAS39505.1 JAS43232.1 JAS47676.1 JAS51910.1 JAS55236.1 JAS57675.1 JAS68033.1 JAS68692.1 GECU01012839 JAS94867.1 NEVH01016298 PNF25914.1 PYGN01001566 PSN34011.1 KK854113 PTY11496.1 KK852601 KDR20513.1 KY285051 ATP16154.1 KQ435922 KOX68475.1 ADTU01013251 KQ976401 KYM92525.1 AK417834 BAN21049.1 KK107522 QOIP01000001 EZA49316.1 RLU27013.1 KQ979851 KYN18846.1 GL888182 EGI65663.1 KQ981490 KYN41074.1 KQ460367 KPJ15459.1 KQ759817 OAD62768.1 KZ288198 PBC33658.1 JR050190 AEY61195.1 KQ978023 KYM97871.1 KQ434998 KZC13334.1 KQ982696 KYQ51978.1 GL453586 EFN75831.1 KQ414582 KOC70892.1 GBHO01017512 JAG26092.1 GEZM01096352 JAV54959.1 GBRD01015156 JAG50670.1 KB632404 ERL95070.1 GECL01003602 JAP02522.1 GDKW01000486 JAI56109.1 LBMM01000231 KMR03596.1 GBGD01000263 JAC88626.1 GBYB01011196 JAG80963.1 LJIG01022490 KRT80302.1 GFTR01008495 JAW07931.1 ACPB03007889 APGK01052863 KB741216 ENN72491.1 GANO01001085 JAB58786.1 GEDC01017015 JAS20283.1 GFDF01004427 JAV09657.1 GFDL01011392 JAV23653.1 GFDL01011285 JAV23760.1 GFDL01011341 JAV23704.1 GFDL01011291 JAV23754.1 GFDL01011325 JAV23720.1 GGMR01004437 MBY17056.1 DS231828 EDS29801.1 GAPW01000070 JAC13528.1 GDIQ01102522 JAL49204.1 LRGB01003241 KZS03606.1 GFXV01001657 MBW13462.1 AXCN02001090 KU998261 ANZ80593.1 ABLF02020541 ABLF02020542 CH477201 EAT48169.1 GDRN01069228 JAI64082.1 GGFJ01001216 MBW50357.1 GGFJ01001217 MBW50358.1 APCN01002415 GGMS01003248 MBY72451.1 EAT48168.1 JQ061292 AFK65746.1 GGFM01006495 MBW27246.1 GGFM01000524 MBW21275.1 GGFM01000526 MBW21277.1 GL732523 EFX89910.1 GEDV01003632 JAP84925.1 AAAB01008986 GACK01005310 JAA59724.1 GDRN01069227 JAI64083.1 GFAC01005767 JAT93421.1 GGFK01000770 MBW34091.1 ADMH02002004 ETN60012.1 GGFK01000752 MBW34073.1 GGFL01003357 MBW67535.1 GBBL01000915 JAC26405.1 MG452698 AYD41585.1 JO842921 AEO34538.1 GEFM01006494 JAP69302.1 GEFH01004093 JAP64488.1
RVE45758.1 KQ459595 KPI95780.1 GAIX01004505 JAA88055.1 AGBW02008504 OWR53156.1 KQ971319 KYB28908.1 GECZ01031811 GECZ01030264 GECZ01026537 GECZ01022093 GECZ01017859 GECZ01014533 GECZ01012094 GECZ01001736 GECZ01001077 JAS37958.1 JAS39505.1 JAS43232.1 JAS47676.1 JAS51910.1 JAS55236.1 JAS57675.1 JAS68033.1 JAS68692.1 GECU01012839 JAS94867.1 NEVH01016298 PNF25914.1 PYGN01001566 PSN34011.1 KK854113 PTY11496.1 KK852601 KDR20513.1 KY285051 ATP16154.1 KQ435922 KOX68475.1 ADTU01013251 KQ976401 KYM92525.1 AK417834 BAN21049.1 KK107522 QOIP01000001 EZA49316.1 RLU27013.1 KQ979851 KYN18846.1 GL888182 EGI65663.1 KQ981490 KYN41074.1 KQ460367 KPJ15459.1 KQ759817 OAD62768.1 KZ288198 PBC33658.1 JR050190 AEY61195.1 KQ978023 KYM97871.1 KQ434998 KZC13334.1 KQ982696 KYQ51978.1 GL453586 EFN75831.1 KQ414582 KOC70892.1 GBHO01017512 JAG26092.1 GEZM01096352 JAV54959.1 GBRD01015156 JAG50670.1 KB632404 ERL95070.1 GECL01003602 JAP02522.1 GDKW01000486 JAI56109.1 LBMM01000231 KMR03596.1 GBGD01000263 JAC88626.1 GBYB01011196 JAG80963.1 LJIG01022490 KRT80302.1 GFTR01008495 JAW07931.1 ACPB03007889 APGK01052863 KB741216 ENN72491.1 GANO01001085 JAB58786.1 GEDC01017015 JAS20283.1 GFDF01004427 JAV09657.1 GFDL01011392 JAV23653.1 GFDL01011285 JAV23760.1 GFDL01011341 JAV23704.1 GFDL01011291 JAV23754.1 GFDL01011325 JAV23720.1 GGMR01004437 MBY17056.1 DS231828 EDS29801.1 GAPW01000070 JAC13528.1 GDIQ01102522 JAL49204.1 LRGB01003241 KZS03606.1 GFXV01001657 MBW13462.1 AXCN02001090 KU998261 ANZ80593.1 ABLF02020541 ABLF02020542 CH477201 EAT48169.1 GDRN01069228 JAI64082.1 GGFJ01001216 MBW50357.1 GGFJ01001217 MBW50358.1 APCN01002415 GGMS01003248 MBY72451.1 EAT48168.1 JQ061292 AFK65746.1 GGFM01006495 MBW27246.1 GGFM01000524 MBW21275.1 GGFM01000526 MBW21277.1 GL732523 EFX89910.1 GEDV01003632 JAP84925.1 AAAB01008986 GACK01005310 JAA59724.1 GDRN01069227 JAI64083.1 GFAC01005767 JAT93421.1 GGFK01000770 MBW34091.1 ADMH02002004 ETN60012.1 GGFK01000752 MBW34073.1 GGFL01003357 MBW67535.1 GBBL01000915 JAC26405.1 MG452698 AYD41585.1 JO842921 AEO34538.1 GEFM01006494 JAP69302.1 GEFH01004093 JAP64488.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000007266
+ More
UP000235965 UP000245037 UP000027135 UP000053105 UP000005205 UP000078540 UP000053097 UP000279307 UP000078492 UP000007755 UP000078541 UP000192223 UP000053240 UP000005203 UP000242457 UP000078542 UP000076502 UP000075809 UP000008237 UP000053825 UP000030742 UP000036403 UP000015103 UP000019118 UP000002320 UP000076858 UP000075886 UP000007819 UP000008820 UP000075881 UP000075840 UP000076407 UP000075903 UP000076408 UP000000305 UP000075885 UP000075880 UP000000673 UP000075900 UP000075884 UP000069272
UP000235965 UP000245037 UP000027135 UP000053105 UP000005205 UP000078540 UP000053097 UP000279307 UP000078492 UP000007755 UP000078541 UP000192223 UP000053240 UP000005203 UP000242457 UP000078542 UP000076502 UP000075809 UP000008237 UP000053825 UP000030742 UP000036403 UP000015103 UP000019118 UP000002320 UP000076858 UP000075886 UP000007819 UP000008820 UP000075881 UP000075840 UP000076407 UP000075903 UP000076408 UP000000305 UP000075885 UP000075880 UP000000673 UP000075900 UP000075884 UP000069272
PRIDE
Pfam
Interpro
IPR035985
Ubiquitin-activating_enz
+ More
IPR018075 UBQ-activ_enz_E1
IPR000594 ThiF_NAD_FAD-bd
IPR038252 UBA_E1_C_sf
IPR000011 UBQ/SUMO-activ_enz_E1-like
IPR018965 Ub-activating_enz_E1_C
IPR019572 UBA_E1_Cys
IPR042302 E1_FCCH_sf
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR032418 E1_FCCH
IPR042063 Ubi_acti_E1_SCCH
IPR018074 UBQ-activ_enz_E1_CS
IPR032420 E1_4HB
IPR016059 DNA_ligase_ATP-dep_CS
IPR012340 NA-bd_OB-fold
IPR012310 DNA_ligase_ATP-dep_cent
IPR016482 SecG/Sec61-beta/Sbh
IPR000977 DNA_ligase_ATP-dep
IPR012308 DNA_ligase_ATP-dep_N
IPR036599 DNA_ligase_N_sf
IPR012309 DNA_ligase_ATP-dep_C
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR019787 Znf_PHD-finger
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR011011 Znf_FYVE_PHD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR018075 UBQ-activ_enz_E1
IPR000594 ThiF_NAD_FAD-bd
IPR038252 UBA_E1_C_sf
IPR000011 UBQ/SUMO-activ_enz_E1-like
IPR018965 Ub-activating_enz_E1_C
IPR019572 UBA_E1_Cys
IPR042302 E1_FCCH_sf
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR032418 E1_FCCH
IPR042063 Ubi_acti_E1_SCCH
IPR018074 UBQ-activ_enz_E1_CS
IPR032420 E1_4HB
IPR016059 DNA_ligase_ATP-dep_CS
IPR012340 NA-bd_OB-fold
IPR012310 DNA_ligase_ATP-dep_cent
IPR016482 SecG/Sec61-beta/Sbh
IPR000977 DNA_ligase_ATP-dep
IPR012308 DNA_ligase_ATP-dep_N
IPR036599 DNA_ligase_N_sf
IPR012309 DNA_ligase_ATP-dep_C
IPR027417 P-loop_NTPase
IPR019821 Kinesin_motor_CS
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR019787 Znf_PHD-finger
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR011011 Znf_FYVE_PHD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR019786 Zinc_finger_PHD-type_CS
SUPFAM
ProteinModelPortal
H9JK60
A0A2H1X2R9
A0A2A4K8C9
A0A3S2PA79
A0A194PXA9
S4PKG8
+ More
A0A212FHF4 A0A139WLH6 A0A1B6EJ56 A0A1B6J6R3 A0A2J7QBF8 A0A2P8XPS9 A0A2R7VVH7 A0A067RA93 A0A2D1QUE3 A0A0M8ZPK0 A0A158NEC8 A0A195BW67 R4WDT6 A0A026W156 A0A151J6R2 F4WJ85 A0A195FM65 A0A1W4WLZ4 A0A194RDB5 A0A1W4WBV3 A0A088AVS9 A0A310SJ05 A0A2A3EPK1 V9IJG1 A0A195CCB1 A0A154PPG8 A0A151WVW5 E2C890 A0A0L7RIX0 A0A0A9XZ86 A0A1Y1K0A8 A0A0K8SD12 U4UXQ1 A0A0V0G561 A0A0P4VU82 A0A0J7P381 A0A069DXS0 A0A0C9RUB8 A0A0T6AZP6 A0A224XIR4 T1HIR2 N6TVS5 U5EX66 A0A1B6D3R9 A0A1L8DTD2 A0A1Q3F816 A0A1Q3F861 A0A1Q3F804 A0A1Q3F854 A0A1Q3F820 A0A2S2NJW1 B0W2P8 A0A023EWP6 A0A0P5RDB7 A0A164KX14 A0A2H8TID4 A0A182QD44 A0A1B2JLV6 J9JZV9 Q17N85 A0A0P4WEI8 A0A182JRA0 A0A2M4BBE8 A0A2M4BC15 A0A2C9GQF2 A0A2S2Q4C1 Q17N86 M9NPG8 A0A2M3ZFB7 A0A182XJW3 A0A182UP22 A0A2M3YYE4 A0A182Y9D9 A0A2M3YYE8 E9FRU9 A0A131Z2F0 A0A1S4H7Q4 L7M9E9 A0A0P4WA61 A0A182PKF3 A0A1E1X320 A0A182JD16 A0A2M4A016 W5J6E3 A0A2M3ZZU6 A0A2M4CQJ6 A0A182RRY5 A0A023G0D6 A0A386H7J3 G3MM20 A0A1Y9H2R6 A0A131XSJ1 A0A131XEH3 A0A182FBD8
A0A212FHF4 A0A139WLH6 A0A1B6EJ56 A0A1B6J6R3 A0A2J7QBF8 A0A2P8XPS9 A0A2R7VVH7 A0A067RA93 A0A2D1QUE3 A0A0M8ZPK0 A0A158NEC8 A0A195BW67 R4WDT6 A0A026W156 A0A151J6R2 F4WJ85 A0A195FM65 A0A1W4WLZ4 A0A194RDB5 A0A1W4WBV3 A0A088AVS9 A0A310SJ05 A0A2A3EPK1 V9IJG1 A0A195CCB1 A0A154PPG8 A0A151WVW5 E2C890 A0A0L7RIX0 A0A0A9XZ86 A0A1Y1K0A8 A0A0K8SD12 U4UXQ1 A0A0V0G561 A0A0P4VU82 A0A0J7P381 A0A069DXS0 A0A0C9RUB8 A0A0T6AZP6 A0A224XIR4 T1HIR2 N6TVS5 U5EX66 A0A1B6D3R9 A0A1L8DTD2 A0A1Q3F816 A0A1Q3F861 A0A1Q3F804 A0A1Q3F854 A0A1Q3F820 A0A2S2NJW1 B0W2P8 A0A023EWP6 A0A0P5RDB7 A0A164KX14 A0A2H8TID4 A0A182QD44 A0A1B2JLV6 J9JZV9 Q17N85 A0A0P4WEI8 A0A182JRA0 A0A2M4BBE8 A0A2M4BC15 A0A2C9GQF2 A0A2S2Q4C1 Q17N86 M9NPG8 A0A2M3ZFB7 A0A182XJW3 A0A182UP22 A0A2M3YYE4 A0A182Y9D9 A0A2M3YYE8 E9FRU9 A0A131Z2F0 A0A1S4H7Q4 L7M9E9 A0A0P4WA61 A0A182PKF3 A0A1E1X320 A0A182JD16 A0A2M4A016 W5J6E3 A0A2M3ZZU6 A0A2M4CQJ6 A0A182RRY5 A0A023G0D6 A0A386H7J3 G3MM20 A0A1Y9H2R6 A0A131XSJ1 A0A131XEH3 A0A182FBD8
PDB
6DC6
E-value=0,
Score=3611
Ontologies
GO
GO:0005524
GO:0008641
GO:0006464
GO:0016874
GO:0005634
GO:0005737
GO:0016567
GO:0006974
GO:0004839
GO:0032446
GO:0016021
GO:0003677
GO:0006260
GO:0071897
GO:0006310
GO:0051103
GO:0003910
GO:0007018
GO:0003777
GO:0008017
GO:0003676
GO:0046872
GO:0016881
GO:0006000
GO:0006003
GO:0006032
GO:0005515
GO:0016742
GO:0003824
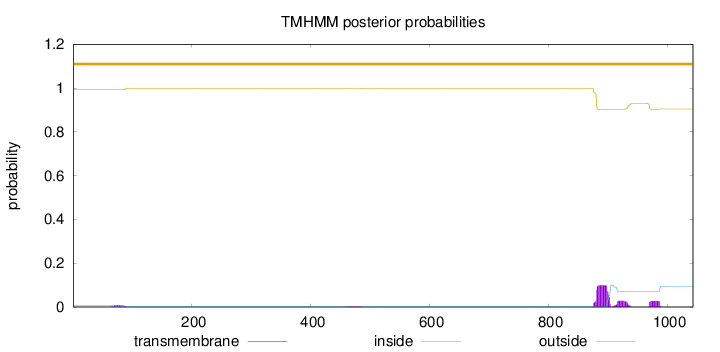
Topology
Length:
1043
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.39845
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.00779
outside
1 - 1043
Population Genetic Test Statistics
Pi
24.878868
Theta
22.273997
Tajima's D
0.260587
CLR
0.639903
CSRT
0.450127493625319
Interpretation
Uncertain
