Gene
KWMTBOMO04803
Pre Gene Modal
BGIBMGA009839
Annotation
PREDICTED:_protein_singed_wings_2_isoform_X2_[Amyelois_transitella]
Full name
Protein halfway
+ More
XK-related protein
XK-related protein
Alternative Name
Protein singed wings
Location in the cell
PlasmaMembrane Reliability : 1.684
Sequence
CDS
ATGTATCTAGCGACATTCCTGCCGTGCATCGAGATGCGGGTTGATGACTTCGACCAGCCCGCACCCAGAGAGCAACCTCCACCGCCGACAGTTCTCCCGGAGCCGATAGAACAGTACTTAAACCTGGGAATATGCTTCCATATGAATCGTTCGTCTTGCCCGCCGAAGGGCGTCTGCACGACCCTAGGGCCCGGCAACATGTCCGTCTTCTGCTGCGATATGGACACTAACTTCTTGAAGGAGTTTCTGCCTGGTATATTGAAACAGAATAAGACGCATCTACACGTGCTGAATGCGACCATCGATGAGCTGGACGTCTCCCACTCCATGTTCAGAAGGCTTACGTCAATGGCGTTTACCGACGGAAATATCAGTAAAATTATTGGTCAGTTCCCGAAGCACTCAGCGCTGGCCTGTTTCAACATATCCAACAACAACTTGACGAGCGACAAGCTGCAGTCTTCGATTCAGAGGCCGTTCGCGTATCTCTTTAACCTGACGGTCCTGGATGCCTCAGCGAACAACCTCACAAAGTTCCCGCTGAGTTTGGTCCACAGCAATAGGAACATATCTGTGGATTTGGCAGGTAACAACTATCTGCCGTGCAAACACTTCCAGAAAGTGATGGAGACGACCAACAGCTCTCTTGTTCACTTCCTCCAGTACAACAAGACCTTCTGCGCTTTGGATCTCAGCTTCAACTGGTTCAAAGACGTCAGCTTCGTGAACATAGACCTTATTAAAATACAGAAAATGATGAATGAGAGATGCCGCGAGATAATACCAGCTGACATCAACTGCACCTGCGCCCCGGAGCGCCTGGAGATCATCGAGAACGAGGTCACCAACCTGGTCTCGGTGGACTGCGCCGAGAGGCAGCTCAAGGAGATGCCGACAAATCTGCCGCCCAATACTGTCAAGTTGAACGTGTCTTTTAATAATATAACATCTCTTGAGGCGGTCGGCGCGGATGAGAGCTACGGACATCTGCGGCAGCTGATCGTGGACTACAACGACATTCCGAATATTCTAGAACTCGAGGGAACGAAGTTCATCGGGAATTTCATGCTCTTCTCGATCGCACATAACAAACTGAAGACTATTCATACGTACGTCTTGTCAAATAGTTTTGAAACGACCGGACCTGCTTTGTCTATAGCGGGAAACTTCATTCACTGCGACTGCAACACCGAGAAGACATTGAAGCCATGGCTATTAGAAAATTTCAAGAGCGTACCGGATTACAAAAGCCTGGAATGCCAGGATGGGCTAGGTCCGGTGGTGGACCTGGTGGAATCTCGCGTCTGCCACACGCCCAGAGATTGGACAGACTACATTTACTACATTATAGGCCTAGAAGTGTTAGTTTTAGTGCTTCTAATCAGCAAAGTATCTTATGACTATTGGGTTTTCAAGACGGCTGGTTATTTGCCGTGGCCTGCGAATAAAATGCCGAGGTTGCCGTGTGATTGGCTCTGCGAGTGA
Protein
MYLATFLPCIEMRVDDFDQPAPREQPPPPTVLPEPIEQYLNLGICFHMNRSSCPPKGVCTTLGPGNMSVFCCDMDTNFLKEFLPGILKQNKTHLHVLNATIDELDVSHSMFRRLTSMAFTDGNISKIIGQFPKHSALACFNISNNNLTSDKLQSSIQRPFAYLFNLTVLDASANNLTKFPLSLVHSNRNISVDLAGNNYLPCKHFQKVMETTNSSLVHFLQYNKTFCALDLSFNWFKDVSFVNIDLIKIQKMMNERCREIIPADINCTCAPERLEIIENEVTNLVSVDCAERQLKEMPTNLPPNTVKLNVSFNNITSLEAVGADESYGHLRQLIVDYNDIPNILELEGTKFIGNFMLFSIAHNKLKTIHTYVLSNSFETTGPALSIAGNFIHCDCNTEKTLKPWLLENFKSVPDYKSLECQDGLGPVVDLVESRVCHTPRDWTDYIYYIIGLEVLVLVLLISKVSYDYWVFKTAGYLPWPANKMPRLPCDWLCE
Summary
Description
Has a role in the ecdysone induced cascade; probably indirect control of 'late' ecdysone genes.
Similarity
Belongs to the XK family.
Keywords
Complete proteome
Glycoprotein
Leucine-rich repeat
Reference proteome
Repeat
Signal
Feature
chain Protein halfway
Uniprot
H9JJY8
A0A194PQQ2
A0A2A4K2Q9
A0A194RC94
A0A212FHE2
A0A0L7LH14
+ More
A0A1Y1L4U5 A0A154NYM4 A0A0L7QS60 A0A088A3M8 A0A026WVP7 D6WG70 A0A224XPH9 A0A2A3E3B6 F4WY96 A0A195CBK6 A0A0J7LBB5 A0A158NHT1 E2AZQ3 N6U0I1 A0A151WR23 E9JB79 A0A195AWR4 A0A0C9RWF5 A0A195FEM7 E2B7P6 A0A1L8DLN0 A0A1L8DLB6 K7J9I7 P84329 A0A0R3P373 A0A195EGB8 A0A0K8UZE7 A0A0K8W957 A0A034V515 R4G331 A0A3B0KJT8 A0A1B0CQ16 B4NCS9 B4PY46 B6IDK2 Q9W568 W8B2N7 B3MYL4 Q16Q64 B3P9K6 A0A1W4VSJ5 A0A2K8JMM7 A0A0L0C549 A0A2S2Q2U7 B4JLV9 A0A067RSI5 A0A2J7Q357 A0A1I8MN61 T1PH04 B4MB41 B4GTJ2 A0A0M4F7J0 A0A182GP64 J9KP37 A0A2H8TTP1 W5JNE4 A0A2M3Z3L0 A0A1I8PL79 A0A1B6DF92 A0A2S2PCE1 A0A2M4BHT4 A0A2M4BH40 A0A2M4BH32 A0A182WW52 A0A182U7C5 A0A182IA38 A0A2M4AS55 Q7QFP4 A0A2M4ASB7 A0A0A9Z1P9 A0A182UZW7 A0A182RES2 A0A182JVS7 A0A182W4E0 A0A182Y7M7 A0A084VV31 A0A182P1N5 A0A182N060 A0A182QDG3 A0A1A9ZLN7 R4WJ71 A0A1B0FI07 A0A1A9UUZ2 A0A182FJS9 A0A182J881 B4L770 A0A0Q9XPZ9 A0A182M327 A0A0M9A889 F0JAN4
A0A1Y1L4U5 A0A154NYM4 A0A0L7QS60 A0A088A3M8 A0A026WVP7 D6WG70 A0A224XPH9 A0A2A3E3B6 F4WY96 A0A195CBK6 A0A0J7LBB5 A0A158NHT1 E2AZQ3 N6U0I1 A0A151WR23 E9JB79 A0A195AWR4 A0A0C9RWF5 A0A195FEM7 E2B7P6 A0A1L8DLN0 A0A1L8DLB6 K7J9I7 P84329 A0A0R3P373 A0A195EGB8 A0A0K8UZE7 A0A0K8W957 A0A034V515 R4G331 A0A3B0KJT8 A0A1B0CQ16 B4NCS9 B4PY46 B6IDK2 Q9W568 W8B2N7 B3MYL4 Q16Q64 B3P9K6 A0A1W4VSJ5 A0A2K8JMM7 A0A0L0C549 A0A2S2Q2U7 B4JLV9 A0A067RSI5 A0A2J7Q357 A0A1I8MN61 T1PH04 B4MB41 B4GTJ2 A0A0M4F7J0 A0A182GP64 J9KP37 A0A2H8TTP1 W5JNE4 A0A2M3Z3L0 A0A1I8PL79 A0A1B6DF92 A0A2S2PCE1 A0A2M4BHT4 A0A2M4BH40 A0A2M4BH32 A0A182WW52 A0A182U7C5 A0A182IA38 A0A2M4AS55 Q7QFP4 A0A2M4ASB7 A0A0A9Z1P9 A0A182UZW7 A0A182RES2 A0A182JVS7 A0A182W4E0 A0A182Y7M7 A0A084VV31 A0A182P1N5 A0A182N060 A0A182QDG3 A0A1A9ZLN7 R4WJ71 A0A1B0FI07 A0A1A9UUZ2 A0A182FJS9 A0A182J881 B4L770 A0A0Q9XPZ9 A0A182M327 A0A0M9A889 F0JAN4
Pubmed
19121390
26354079
22118469
26227816
28004739
24508170
+ More
30249741 18362917 19820115 21719571 21347285 20798317 23537049 21282665 20075255 15632085 15189568 17994087 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10731137 12537569 17893096 19349973 24495485 17510324 26108605 24845553 25315136 26483478 20920257 23761445 12364791 14747013 17210077 25401762 26823975 25244985 24438588 23691247
30249741 18362917 19820115 21719571 21347285 20798317 23537049 21282665 20075255 15632085 15189568 17994087 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10731137 12537569 17893096 19349973 24495485 17510324 26108605 24845553 25315136 26483478 20920257 23761445 12364791 14747013 17210077 25401762 26823975 25244985 24438588 23691247
EMBL
BABH01026699
BABH01026700
KQ459595
KPI95776.1
NWSH01000260
PCG77932.1
+ More
KQ460367 KPJ15463.1 AGBW02008504 OWR53160.1 JTDY01001177 KOB74644.1 GEZM01064590 JAV68703.1 KQ434783 KZC04723.1 KQ414758 KOC61472.1 KK107109 QOIP01000008 EZA59164.1 RLU19156.1 KQ971320 EFA00539.1 GFTR01006495 JAW09931.1 KZ288403 PBC26217.1 GL888439 EGI60853.1 KQ978023 KYM98080.1 LBMM01000040 KMR05202.1 ADTU01016037 GL444277 EFN61032.1 APGK01047050 KB741077 KB632384 ENN74081.1 ERL94212.1 KQ982813 KYQ50258.1 GL770938 EFZ09931.1 KQ976725 KYM76611.1 GBYB01011310 GBYB01012126 GBYB01012127 JAG81077.1 JAG81893.1 JAG81894.1 KQ981636 KYN38853.1 GL446201 EFN88259.1 GFDF01006820 JAV07264.1 GFDF01006821 JAV07263.1 AAZX01012840 AAZX01019089 CH379063 KRT06077.1 KQ978957 KYN27283.1 GDHF01020267 JAI32047.1 GDHF01004929 JAI47385.1 GAKP01020571 JAC38381.1 ACPB03020356 GAHY01002046 JAA75464.1 OUUW01000011 SPP86759.1 AJWK01022922 CH964239 EDW82638.2 CM000162 EDX00919.1 BT050442 AE014298 ACJ13149.1 AGB94981.1 AJ626646 AL021726 AY070931 AAF45654.3 AAL48553.1 CAA16810.1 CAF25166.1 GAMC01015252 JAB91303.1 CH902632 EDV32708.1 CH477759 EAT36542.1 CH954183 EDV45502.1 KY031173 ATU82924.1 JRES01000902 KNC27395.1 GGMS01002777 MBY71980.1 CH916371 EDV91720.1 KK852488 KDR22774.1 NEVH01019069 PNF23031.1 KA648046 AFP62675.1 CH940655 EDW66450.2 CH479190 EDW25862.1 CP012528 ALC48175.1 JXUM01014868 JXUM01014869 JXUM01014870 JXUM01014871 JXUM01014872 KQ560415 KXJ82525.1 ABLF02011290 ABLF02035754 ABLF02063648 GFXV01005772 MBW17577.1 ADMH02000558 ETN65887.1 GGFM01002360 MBW23111.1 GEDC01020848 GEDC01012951 JAS16450.1 JAS24347.1 GGMR01014239 MBY26858.1 GGFJ01003227 MBW52368.1 GGFJ01003229 MBW52370.1 GGFJ01003228 MBW52369.1 APCN01001102 GGFK01010315 MBW43636.1 AAAB01008846 EAA06520.4 EGK96403.1 GGFK01010339 MBW43660.1 GBHO01038385 GBHO01038384 GBHO01038383 GBHO01038381 GBHO01007874 GBRD01001069 GDHC01012491 JAG05219.1 JAG05220.1 JAG05221.1 JAG05223.1 JAG35730.1 JAG64752.1 JAQ06138.1 ATLV01017149 KE525157 KFB41825.1 AXCN02001308 AK417525 BAN20740.1 CCAG010009519 AXCP01008108 CH933812 EDW06216.2 KRG07186.1 AXCM01001777 KQ435724 KOX78486.1 BT126076 ADY17775.1
KQ460367 KPJ15463.1 AGBW02008504 OWR53160.1 JTDY01001177 KOB74644.1 GEZM01064590 JAV68703.1 KQ434783 KZC04723.1 KQ414758 KOC61472.1 KK107109 QOIP01000008 EZA59164.1 RLU19156.1 KQ971320 EFA00539.1 GFTR01006495 JAW09931.1 KZ288403 PBC26217.1 GL888439 EGI60853.1 KQ978023 KYM98080.1 LBMM01000040 KMR05202.1 ADTU01016037 GL444277 EFN61032.1 APGK01047050 KB741077 KB632384 ENN74081.1 ERL94212.1 KQ982813 KYQ50258.1 GL770938 EFZ09931.1 KQ976725 KYM76611.1 GBYB01011310 GBYB01012126 GBYB01012127 JAG81077.1 JAG81893.1 JAG81894.1 KQ981636 KYN38853.1 GL446201 EFN88259.1 GFDF01006820 JAV07264.1 GFDF01006821 JAV07263.1 AAZX01012840 AAZX01019089 CH379063 KRT06077.1 KQ978957 KYN27283.1 GDHF01020267 JAI32047.1 GDHF01004929 JAI47385.1 GAKP01020571 JAC38381.1 ACPB03020356 GAHY01002046 JAA75464.1 OUUW01000011 SPP86759.1 AJWK01022922 CH964239 EDW82638.2 CM000162 EDX00919.1 BT050442 AE014298 ACJ13149.1 AGB94981.1 AJ626646 AL021726 AY070931 AAF45654.3 AAL48553.1 CAA16810.1 CAF25166.1 GAMC01015252 JAB91303.1 CH902632 EDV32708.1 CH477759 EAT36542.1 CH954183 EDV45502.1 KY031173 ATU82924.1 JRES01000902 KNC27395.1 GGMS01002777 MBY71980.1 CH916371 EDV91720.1 KK852488 KDR22774.1 NEVH01019069 PNF23031.1 KA648046 AFP62675.1 CH940655 EDW66450.2 CH479190 EDW25862.1 CP012528 ALC48175.1 JXUM01014868 JXUM01014869 JXUM01014870 JXUM01014871 JXUM01014872 KQ560415 KXJ82525.1 ABLF02011290 ABLF02035754 ABLF02063648 GFXV01005772 MBW17577.1 ADMH02000558 ETN65887.1 GGFM01002360 MBW23111.1 GEDC01020848 GEDC01012951 JAS16450.1 JAS24347.1 GGMR01014239 MBY26858.1 GGFJ01003227 MBW52368.1 GGFJ01003229 MBW52370.1 GGFJ01003228 MBW52369.1 APCN01001102 GGFK01010315 MBW43636.1 AAAB01008846 EAA06520.4 EGK96403.1 GGFK01010339 MBW43660.1 GBHO01038385 GBHO01038384 GBHO01038383 GBHO01038381 GBHO01007874 GBRD01001069 GDHC01012491 JAG05219.1 JAG05220.1 JAG05221.1 JAG05223.1 JAG35730.1 JAG64752.1 JAQ06138.1 ATLV01017149 KE525157 KFB41825.1 AXCN02001308 AK417525 BAN20740.1 CCAG010009519 AXCP01008108 CH933812 EDW06216.2 KRG07186.1 AXCM01001777 KQ435724 KOX78486.1 BT126076 ADY17775.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000037510
+ More
UP000076502 UP000053825 UP000005203 UP000053097 UP000279307 UP000007266 UP000242457 UP000007755 UP000078542 UP000036403 UP000005205 UP000000311 UP000019118 UP000030742 UP000075809 UP000078540 UP000078541 UP000008237 UP000002358 UP000001819 UP000078492 UP000015103 UP000268350 UP000092461 UP000007798 UP000002282 UP000000803 UP000007801 UP000008820 UP000008711 UP000192221 UP000037069 UP000001070 UP000027135 UP000235965 UP000095301 UP000008792 UP000008744 UP000092553 UP000069940 UP000249989 UP000007819 UP000000673 UP000095300 UP000076407 UP000075902 UP000075840 UP000007062 UP000075903 UP000075900 UP000075881 UP000075920 UP000076408 UP000030765 UP000075885 UP000075884 UP000075886 UP000092445 UP000092444 UP000078200 UP000069272 UP000075880 UP000009192 UP000075883 UP000053105
UP000076502 UP000053825 UP000005203 UP000053097 UP000279307 UP000007266 UP000242457 UP000007755 UP000078542 UP000036403 UP000005205 UP000000311 UP000019118 UP000030742 UP000075809 UP000078540 UP000078541 UP000008237 UP000002358 UP000001819 UP000078492 UP000015103 UP000268350 UP000092461 UP000007798 UP000002282 UP000000803 UP000007801 UP000008820 UP000008711 UP000192221 UP000037069 UP000001070 UP000027135 UP000235965 UP000095301 UP000008792 UP000008744 UP000092553 UP000069940 UP000249989 UP000007819 UP000000673 UP000095300 UP000076407 UP000075902 UP000075840 UP000007062 UP000075903 UP000075900 UP000075881 UP000075920 UP000076408 UP000030765 UP000075885 UP000075884 UP000075886 UP000092445 UP000092444 UP000078200 UP000069272 UP000075880 UP000009192 UP000075883 UP000053105
Pfam
Interpro
SUPFAM
SSF69065
SSF69065
Gene 3D
ProteinModelPortal
H9JJY8
A0A194PQQ2
A0A2A4K2Q9
A0A194RC94
A0A212FHE2
A0A0L7LH14
+ More
A0A1Y1L4U5 A0A154NYM4 A0A0L7QS60 A0A088A3M8 A0A026WVP7 D6WG70 A0A224XPH9 A0A2A3E3B6 F4WY96 A0A195CBK6 A0A0J7LBB5 A0A158NHT1 E2AZQ3 N6U0I1 A0A151WR23 E9JB79 A0A195AWR4 A0A0C9RWF5 A0A195FEM7 E2B7P6 A0A1L8DLN0 A0A1L8DLB6 K7J9I7 P84329 A0A0R3P373 A0A195EGB8 A0A0K8UZE7 A0A0K8W957 A0A034V515 R4G331 A0A3B0KJT8 A0A1B0CQ16 B4NCS9 B4PY46 B6IDK2 Q9W568 W8B2N7 B3MYL4 Q16Q64 B3P9K6 A0A1W4VSJ5 A0A2K8JMM7 A0A0L0C549 A0A2S2Q2U7 B4JLV9 A0A067RSI5 A0A2J7Q357 A0A1I8MN61 T1PH04 B4MB41 B4GTJ2 A0A0M4F7J0 A0A182GP64 J9KP37 A0A2H8TTP1 W5JNE4 A0A2M3Z3L0 A0A1I8PL79 A0A1B6DF92 A0A2S2PCE1 A0A2M4BHT4 A0A2M4BH40 A0A2M4BH32 A0A182WW52 A0A182U7C5 A0A182IA38 A0A2M4AS55 Q7QFP4 A0A2M4ASB7 A0A0A9Z1P9 A0A182UZW7 A0A182RES2 A0A182JVS7 A0A182W4E0 A0A182Y7M7 A0A084VV31 A0A182P1N5 A0A182N060 A0A182QDG3 A0A1A9ZLN7 R4WJ71 A0A1B0FI07 A0A1A9UUZ2 A0A182FJS9 A0A182J881 B4L770 A0A0Q9XPZ9 A0A182M327 A0A0M9A889 F0JAN4
A0A1Y1L4U5 A0A154NYM4 A0A0L7QS60 A0A088A3M8 A0A026WVP7 D6WG70 A0A224XPH9 A0A2A3E3B6 F4WY96 A0A195CBK6 A0A0J7LBB5 A0A158NHT1 E2AZQ3 N6U0I1 A0A151WR23 E9JB79 A0A195AWR4 A0A0C9RWF5 A0A195FEM7 E2B7P6 A0A1L8DLN0 A0A1L8DLB6 K7J9I7 P84329 A0A0R3P373 A0A195EGB8 A0A0K8UZE7 A0A0K8W957 A0A034V515 R4G331 A0A3B0KJT8 A0A1B0CQ16 B4NCS9 B4PY46 B6IDK2 Q9W568 W8B2N7 B3MYL4 Q16Q64 B3P9K6 A0A1W4VSJ5 A0A2K8JMM7 A0A0L0C549 A0A2S2Q2U7 B4JLV9 A0A067RSI5 A0A2J7Q357 A0A1I8MN61 T1PH04 B4MB41 B4GTJ2 A0A0M4F7J0 A0A182GP64 J9KP37 A0A2H8TTP1 W5JNE4 A0A2M3Z3L0 A0A1I8PL79 A0A1B6DF92 A0A2S2PCE1 A0A2M4BHT4 A0A2M4BH40 A0A2M4BH32 A0A182WW52 A0A182U7C5 A0A182IA38 A0A2M4AS55 Q7QFP4 A0A2M4ASB7 A0A0A9Z1P9 A0A182UZW7 A0A182RES2 A0A182JVS7 A0A182W4E0 A0A182Y7M7 A0A084VV31 A0A182P1N5 A0A182N060 A0A182QDG3 A0A1A9ZLN7 R4WJ71 A0A1B0FI07 A0A1A9UUZ2 A0A182FJS9 A0A182J881 B4L770 A0A0Q9XPZ9 A0A182M327 A0A0M9A889 F0JAN4
PDB
2XOT
E-value=0.000292106,
Score=106
Ontologies
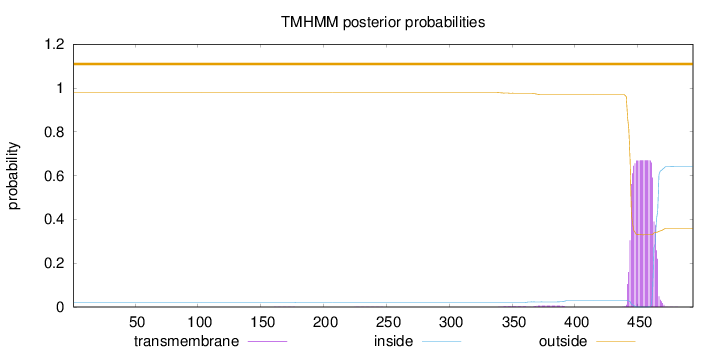
Topology
Subcellular location
Membrane
Length:
494
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.93792
Exp number, first 60 AAs:
4e-05
Total prob of N-in:
0.02019
outside
1 - 494
Population Genetic Test Statistics
Pi
22.445251
Theta
19.729101
Tajima's D
0.378018
CLR
0.582274
CSRT
0.483025848707565
Interpretation
Uncertain
