Pre Gene Modal
BGIBMGA009910
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_DDX52_[Plutella_xylostella]
Full name
Eukaryotic translation initiation factor 3 subunit H
Location in the cell
Nuclear Reliability : 4.26
Sequence
CDS
ATGGACGCATACGATATATTCAAGAAATTAACGAAAGGATTGTCATTTAAAAAACCTGTGTTGGGTGTTAAAAACAATCAACAGAAAACTAAACAGGAGACCTTAAAACAAGAAATTGAAATCAAGAAGGAAGAATTGAGCGACTTTGAAGATGTTGCGGAAATTGAGAATGATTTGGAAAACTCCCAACCAAGTTCTGGAAGCGAGGAAGAAGAATTGGAGCTAGTTGATGGTGTTAAAGTTGGTAAAAAAAAGCGGGGAAAGAAAAAGAAGGAAATAACCGAAGATTTAAAGAAGAAACTGTTACAAGAGGAGGAAAATCGTTTTCGTAATGAACATGGTATCAAAGCAGTCGGCCGACACATACCACCAGCACTAAAAGACTTTTCGGACTTGACTGTCCGTTACAATGTTCCACAGGCTCTTGTTGATACAGTCACCCAGTGTGGATACAGTGAACCCACTCCAGTTCAACGTCAAGCAATGGCTTGCATGTTGGAGGACCGTCAAATAGTAGCATGCGCTCCTACGGGGTCGGGTAAAACTGCCGCGTTTATCGTACCCCTCCTTCATACATTAGGAACGCACCAAGGCGGTCCTAGAGCCCTAATACTGTGCCCTACTAGGGAACTGGCTCATCAAATATACAGGGAGGCCCTACGGCTGAGTGCCTCAACTCAACTTAGAGTTACAGTTGTGAAGAATTTGAAAGAAAGTAAAGTTAAAGAACGCGAAGCTACCTTCCGGAAAAGCGATATAGTTATAAGTACACCAAATCGACTCTGCTATCTACTGAATCAAGATCAAGTTAATATTTCTTTAGAGAAAGTACGCTGGCTTATAATCGACGAGGCCGACAAGCTGTTCGAGGGCTCCTCAGAGCAGGAATCCGACTTCAGGCAGCAGCTGGAGCAGATAATGTGCCGCTGTGGACCGAAACGCCTCATCGCCATGTTCAGCGCCACGCACACGCCGCCCGTCGCCCGCTGGAGCCGCCGCCACCTGCGCGGGCTCGTCAACGTCACCGTGGGACACAGGAACGCTGCGACGAGCCTGGTGGAGCAGCAGCTGCTGTACTGCGGCAACGAGGATGGCAAGCTGGTGGCGTTCCGGCAGCTCGTGCAGCAAGGACTCAAGCCCCCCGTACTAGTCTTCGTACAAAGTAAAGATCGCGCGAAACAATTGTTCAAGGAACTCATATATGACGGCATCAACGTGGACGTCATACACGCAGACAGGACTCAGGCTCAGCGAGACAACGTGGTCCGCAGCTTCCGCGTGGGACGTATATGGGTCCTGATCAGCACCGAACTGATGGGCCGCGGGATCGACTTCCGGGGAGTGAATCTGGTCATCAACTACGACTTCCCACCCTCAGCTATATCTTACGTACACAGGATCGGGCGTGCGGGCCGGGCCGGTCACAGGGGGAGGGCTGTGACGTTCTTCACGCAGGACGACGTCGTCAACTTGAGGAGCATCGCGTCGGTGATGAAGCAGTCCGGCTGCGAGGTCCCGGAGTACATGCTGAGGCTGAAGCAAGACTCCAACAAGCGAAAACGGCTGCTCAAGAAGGCTCCGCATCGAGATAAAATATCCACTATACTAGAGAAGCCGGTCAAAAGGAAATTAGAAGAAGGAAAGGATTCTAATGTAAATTCAAAAGATACAGAGAAGATAAAGAACGGTAAACACAAGAAGCAAAGGAAAATTGATGGAACAAAAGAAAATGGCGCTCGCAGAAGTACGAGCGAGAAGGAGAATAAAAGGAAAAAATTTAAAAAGAATAAAAAGATTAAATTAACCAAAAAACCTAAATCTGGAAGTTGA
Protein
MDAYDIFKKLTKGLSFKKPVLGVKNNQQKTKQETLKQEIEIKKEELSDFEDVAEIENDLENSQPSSGSEEEELELVDGVKVGKKKRGKKKKEITEDLKKKLLQEEENRFRNEHGIKAVGRHIPPALKDFSDLTVRYNVPQALVDTVTQCGYSEPTPVQRQAMACMLEDRQIVACAPTGSGKTAAFIVPLLHTLGTHQGGPRALILCPTRELAHQIYREALRLSASTQLRVTVVKNLKESKVKEREATFRKSDIVISTPNRLCYLLNQDQVNISLEKVRWLIIDEADKLFEGSSEQESDFRQQLEQIMCRCGPKRLIAMFSATHTPPVARWSRRHLRGLVNVTVGHRNAATSLVEQQLLYCGNEDGKLVAFRQLVQQGLKPPVLVFVQSKDRAKQLFKELIYDGINVDVIHADRTQAQRDNVVRSFRVGRIWVLISTELMGRGIDFRGVNLVINYDFPPSAISYVHRIGRAGRAGHRGRAVTFFTQDDVVNLRSIASVMKQSGCEVPEYMLRLKQDSNKRKRLLKKAPHRDKISTILEKPVKRKLEEGKDSNVNSKDTEKIKNGKHKKQRKIDGTKENGARRSTSEKENKRKKFKKNKKIKLTKKPKSGS
Summary
Description
Component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is involved in protein synthesis of a specialized repertoire of mRNAs and, together with other initiation factors, stimulates binding of mRNA and methionyl-tRNAi to the 40S ribosome. The eIF-3 complex specifically targets and initiates translation of a subset of mRNAs involved in cell proliferation.
Subunit
Component of the eukaryotic translation initiation factor 3 (eIF-3) complex.
Similarity
Belongs to the DEAD box helicase family.
Belongs to the eIF-3 subunit H family.
Belongs to the eIF-3 subunit H family.
Uniprot
H9JK59
A0A2A4K0Y6
A0A194PXA4
A0A194RDC1
A0A0L7QSM7
A0A154P053
+ More
A0A2A3E4M6 D6WHF8 A0A026WT93 A0A195CCY7 A0A195EHD4 A0A195FF55 A0A067QRL4 F4WY97 A0A2J7Q1B3 A0A2J7Q1A1 A0A195AX39 A0A158NHT0 A0A1W4XMH0 A0A151WQT2 A0A088A3I3 K7IRY2 A0A232FJM5 A0A1B6CLU8 N6T2V3 A0A2R7W2G6 J3JWN9 A0A1A9X027 A0A1A9UUY7 A0A1B0ASX3 A0A1B0C8R4 A0A1A9XYC7 A0A1B6MTW2 U4UXL9 A0A0L0C588 A0A1B0FI31 B4N4N5 A0A1I8NUX7 A0A1A9ZLN9 A0A2S2PW99 A0A1B6HEI1 A0A1B6FC57 E2BXA2 B3M3V9 B4GZQ4 B5DQ82 B4L0G8 A0A3B0JM30 B4LIG4 A0A1L8DGA2 A0A2H8TT76 A0A1I8MKR7 A0A182GP65 A0A1S4FXZ9 Q16KK0 A0A182P1R6 B3NHR1 A0A0M5JB23 E9GSQ0 A0A0P5WU69 A0A0A9WXQ0 A0A1W4W533 A0A0P4YE59 A0A146MG33 A0A1Y1L408 A0A0P5X025 A0A084WQN8 A0A2B4S2X6 A0A0P5WSG2 A0A0P4ZJU7 B4HKW8 A0A0P5MJ44 A0A0R1DYK2 B4PJJ1 A0A0J9UNE0 A0A224XM11 B4QPA7 Q9VVK8 A0A0P5MBJ4 A0A0P6CP43 A0A0P6F8X0 B5X544 A0A1B0CQ14 H9ZYP2 A0A182V1B8 A0A182RB65 A0A182TKG6 A0A182HQ94
A0A2A3E4M6 D6WHF8 A0A026WT93 A0A195CCY7 A0A195EHD4 A0A195FF55 A0A067QRL4 F4WY97 A0A2J7Q1B3 A0A2J7Q1A1 A0A195AX39 A0A158NHT0 A0A1W4XMH0 A0A151WQT2 A0A088A3I3 K7IRY2 A0A232FJM5 A0A1B6CLU8 N6T2V3 A0A2R7W2G6 J3JWN9 A0A1A9X027 A0A1A9UUY7 A0A1B0ASX3 A0A1B0C8R4 A0A1A9XYC7 A0A1B6MTW2 U4UXL9 A0A0L0C588 A0A1B0FI31 B4N4N5 A0A1I8NUX7 A0A1A9ZLN9 A0A2S2PW99 A0A1B6HEI1 A0A1B6FC57 E2BXA2 B3M3V9 B4GZQ4 B5DQ82 B4L0G8 A0A3B0JM30 B4LIG4 A0A1L8DGA2 A0A2H8TT76 A0A1I8MKR7 A0A182GP65 A0A1S4FXZ9 Q16KK0 A0A182P1R6 B3NHR1 A0A0M5JB23 E9GSQ0 A0A0P5WU69 A0A0A9WXQ0 A0A1W4W533 A0A0P4YE59 A0A146MG33 A0A1Y1L408 A0A0P5X025 A0A084WQN8 A0A2B4S2X6 A0A0P5WSG2 A0A0P4ZJU7 B4HKW8 A0A0P5MJ44 A0A0R1DYK2 B4PJJ1 A0A0J9UNE0 A0A224XM11 B4QPA7 Q9VVK8 A0A0P5MBJ4 A0A0P6CP43 A0A0P6F8X0 B5X544 A0A1B0CQ14 H9ZYP2 A0A182V1B8 A0A182RB65 A0A182TKG6 A0A182HQ94
Pubmed
19121390
26354079
18362917
19820115
24508170
30249741
+ More
24845553 21719571 21347285 20075255 28648823 23537049 22516182 26108605 17994087 20798317 15632085 18057021 25315136 26483478 17510324 21292972 25401762 26823975 28004739 24438588 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
24845553 21719571 21347285 20075255 28648823 23537049 22516182 26108605 17994087 20798317 15632085 18057021 25315136 26483478 17510324 21292972 25401762 26823975 28004739 24438588 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
EMBL
BABH01026699
NWSH01000260
PCG77931.1
KQ459595
KPI95775.1
KQ460367
+ More
KPJ15464.1 KQ414758 KOC61471.1 KQ434783 KZC04724.1 KZ288403 PBC26216.1 KQ971321 EFA00097.2 KK107109 QOIP01000008 EZA59163.1 RLU19646.1 KQ978023 KYM98081.1 KQ978957 KYN27282.1 KQ981636 KYN38852.1 KK853042 KDR12154.1 GL888439 EGI60854.1 NEVH01019430 PNF22367.1 PNF22364.1 KQ976725 KYM76610.1 ADTU01016037 KQ982813 KYQ50259.1 NNAY01000103 OXU30946.1 GEDC01022896 JAS14402.1 APGK01054677 APGK01054678 APGK01054679 APGK01054680 KB741251 ENN71883.1 KK854261 PTY13872.1 BT127657 AEE62619.1 JXJN01003037 AJWK01001283 GEBQ01000648 JAT39329.1 KB632404 ERL95035.1 JRES01000902 KNC27396.1 CCAG010009519 CH964095 EDW79109.2 GGMS01000421 MBY69624.1 GECU01034618 JAS73088.1 GECZ01021972 JAS47797.1 GL451230 EFN79685.1 CH902618 EDV39293.2 CH479199 EDW29481.1 CH379069 EDY73775.1 CH933809 EDW19137.1 OUUW01000002 SPP76480.1 CH940647 EDW70751.1 KRF85084.1 GFDF01008608 JAV05476.1 GFXV01004987 MBW16792.1 JXUM01014872 KQ560415 KXJ82526.1 CH477953 EAT34832.1 CH954178 EDV51926.1 CP012525 ALC43856.1 GL732562 EFX77543.1 GDIQ01188150 GDIP01082045 JAK63575.1 JAM21670.1 GBHO01032291 JAG11313.1 GDIP01232333 JAI91068.1 GDHC01000742 JAQ17887.1 GEZM01068203 JAV67100.1 GDIP01079637 JAM24078.1 ATLV01025642 KE525396 KFB52532.1 LSMT01000209 PFX23393.1 GDIP01082044 JAM21671.1 GDIP01215589 JAJ07813.1 CH480815 EDW41923.1 GDIQ01204251 GDIQ01155869 GDIQ01092694 GDIQ01036256 GDIQ01030283 LRGB01000687 JAK95856.1 KZS16950.1 CM000159 KRK01983.1 EDW94679.1 CM002912 KMZ00311.1 GFTR01006794 JAW09632.1 CM000363 EDX10901.1 AE014296 AAF49303.1 GDIQ01158017 JAK93708.1 GDIQ01090238 JAN04499.1 GDIQ01052321 JAN42416.1 BT046163 ACI46551.1 AJWK01022922 BT133390 AFH41849.1 APCN01003303
KPJ15464.1 KQ414758 KOC61471.1 KQ434783 KZC04724.1 KZ288403 PBC26216.1 KQ971321 EFA00097.2 KK107109 QOIP01000008 EZA59163.1 RLU19646.1 KQ978023 KYM98081.1 KQ978957 KYN27282.1 KQ981636 KYN38852.1 KK853042 KDR12154.1 GL888439 EGI60854.1 NEVH01019430 PNF22367.1 PNF22364.1 KQ976725 KYM76610.1 ADTU01016037 KQ982813 KYQ50259.1 NNAY01000103 OXU30946.1 GEDC01022896 JAS14402.1 APGK01054677 APGK01054678 APGK01054679 APGK01054680 KB741251 ENN71883.1 KK854261 PTY13872.1 BT127657 AEE62619.1 JXJN01003037 AJWK01001283 GEBQ01000648 JAT39329.1 KB632404 ERL95035.1 JRES01000902 KNC27396.1 CCAG010009519 CH964095 EDW79109.2 GGMS01000421 MBY69624.1 GECU01034618 JAS73088.1 GECZ01021972 JAS47797.1 GL451230 EFN79685.1 CH902618 EDV39293.2 CH479199 EDW29481.1 CH379069 EDY73775.1 CH933809 EDW19137.1 OUUW01000002 SPP76480.1 CH940647 EDW70751.1 KRF85084.1 GFDF01008608 JAV05476.1 GFXV01004987 MBW16792.1 JXUM01014872 KQ560415 KXJ82526.1 CH477953 EAT34832.1 CH954178 EDV51926.1 CP012525 ALC43856.1 GL732562 EFX77543.1 GDIQ01188150 GDIP01082045 JAK63575.1 JAM21670.1 GBHO01032291 JAG11313.1 GDIP01232333 JAI91068.1 GDHC01000742 JAQ17887.1 GEZM01068203 JAV67100.1 GDIP01079637 JAM24078.1 ATLV01025642 KE525396 KFB52532.1 LSMT01000209 PFX23393.1 GDIP01082044 JAM21671.1 GDIP01215589 JAJ07813.1 CH480815 EDW41923.1 GDIQ01204251 GDIQ01155869 GDIQ01092694 GDIQ01036256 GDIQ01030283 LRGB01000687 JAK95856.1 KZS16950.1 CM000159 KRK01983.1 EDW94679.1 CM002912 KMZ00311.1 GFTR01006794 JAW09632.1 CM000363 EDX10901.1 AE014296 AAF49303.1 GDIQ01158017 JAK93708.1 GDIQ01090238 JAN04499.1 GDIQ01052321 JAN42416.1 BT046163 ACI46551.1 AJWK01022922 BT133390 AFH41849.1 APCN01003303
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000053825
UP000076502
+ More
UP000242457 UP000007266 UP000053097 UP000279307 UP000078542 UP000078492 UP000078541 UP000027135 UP000007755 UP000235965 UP000078540 UP000005205 UP000192223 UP000075809 UP000005203 UP000002358 UP000215335 UP000019118 UP000091820 UP000078200 UP000092460 UP000092461 UP000092443 UP000030742 UP000037069 UP000092444 UP000007798 UP000095300 UP000092445 UP000008237 UP000007801 UP000008744 UP000001819 UP000009192 UP000268350 UP000008792 UP000095301 UP000069940 UP000249989 UP000008820 UP000075885 UP000008711 UP000092553 UP000000305 UP000192221 UP000030765 UP000225706 UP000001292 UP000076858 UP000002282 UP000000304 UP000000803 UP000075903 UP000075900 UP000075902 UP000075840
UP000242457 UP000007266 UP000053097 UP000279307 UP000078542 UP000078492 UP000078541 UP000027135 UP000007755 UP000235965 UP000078540 UP000005205 UP000192223 UP000075809 UP000005203 UP000002358 UP000215335 UP000019118 UP000091820 UP000078200 UP000092460 UP000092461 UP000092443 UP000030742 UP000037069 UP000092444 UP000007798 UP000095300 UP000092445 UP000008237 UP000007801 UP000008744 UP000001819 UP000009192 UP000268350 UP000008792 UP000095301 UP000069940 UP000249989 UP000008820 UP000075885 UP000008711 UP000092553 UP000000305 UP000192221 UP000030765 UP000225706 UP000001292 UP000076858 UP000002282 UP000000304 UP000000803 UP000075903 UP000075900 UP000075902 UP000075840
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JK59
A0A2A4K0Y6
A0A194PXA4
A0A194RDC1
A0A0L7QSM7
A0A154P053
+ More
A0A2A3E4M6 D6WHF8 A0A026WT93 A0A195CCY7 A0A195EHD4 A0A195FF55 A0A067QRL4 F4WY97 A0A2J7Q1B3 A0A2J7Q1A1 A0A195AX39 A0A158NHT0 A0A1W4XMH0 A0A151WQT2 A0A088A3I3 K7IRY2 A0A232FJM5 A0A1B6CLU8 N6T2V3 A0A2R7W2G6 J3JWN9 A0A1A9X027 A0A1A9UUY7 A0A1B0ASX3 A0A1B0C8R4 A0A1A9XYC7 A0A1B6MTW2 U4UXL9 A0A0L0C588 A0A1B0FI31 B4N4N5 A0A1I8NUX7 A0A1A9ZLN9 A0A2S2PW99 A0A1B6HEI1 A0A1B6FC57 E2BXA2 B3M3V9 B4GZQ4 B5DQ82 B4L0G8 A0A3B0JM30 B4LIG4 A0A1L8DGA2 A0A2H8TT76 A0A1I8MKR7 A0A182GP65 A0A1S4FXZ9 Q16KK0 A0A182P1R6 B3NHR1 A0A0M5JB23 E9GSQ0 A0A0P5WU69 A0A0A9WXQ0 A0A1W4W533 A0A0P4YE59 A0A146MG33 A0A1Y1L408 A0A0P5X025 A0A084WQN8 A0A2B4S2X6 A0A0P5WSG2 A0A0P4ZJU7 B4HKW8 A0A0P5MJ44 A0A0R1DYK2 B4PJJ1 A0A0J9UNE0 A0A224XM11 B4QPA7 Q9VVK8 A0A0P5MBJ4 A0A0P6CP43 A0A0P6F8X0 B5X544 A0A1B0CQ14 H9ZYP2 A0A182V1B8 A0A182RB65 A0A182TKG6 A0A182HQ94
A0A2A3E4M6 D6WHF8 A0A026WT93 A0A195CCY7 A0A195EHD4 A0A195FF55 A0A067QRL4 F4WY97 A0A2J7Q1B3 A0A2J7Q1A1 A0A195AX39 A0A158NHT0 A0A1W4XMH0 A0A151WQT2 A0A088A3I3 K7IRY2 A0A232FJM5 A0A1B6CLU8 N6T2V3 A0A2R7W2G6 J3JWN9 A0A1A9X027 A0A1A9UUY7 A0A1B0ASX3 A0A1B0C8R4 A0A1A9XYC7 A0A1B6MTW2 U4UXL9 A0A0L0C588 A0A1B0FI31 B4N4N5 A0A1I8NUX7 A0A1A9ZLN9 A0A2S2PW99 A0A1B6HEI1 A0A1B6FC57 E2BXA2 B3M3V9 B4GZQ4 B5DQ82 B4L0G8 A0A3B0JM30 B4LIG4 A0A1L8DGA2 A0A2H8TT76 A0A1I8MKR7 A0A182GP65 A0A1S4FXZ9 Q16KK0 A0A182P1R6 B3NHR1 A0A0M5JB23 E9GSQ0 A0A0P5WU69 A0A0A9WXQ0 A0A1W4W533 A0A0P4YE59 A0A146MG33 A0A1Y1L408 A0A0P5X025 A0A084WQN8 A0A2B4S2X6 A0A0P5WSG2 A0A0P4ZJU7 B4HKW8 A0A0P5MJ44 A0A0R1DYK2 B4PJJ1 A0A0J9UNE0 A0A224XM11 B4QPA7 Q9VVK8 A0A0P5MBJ4 A0A0P6CP43 A0A0P6F8X0 B5X544 A0A1B0CQ14 H9ZYP2 A0A182V1B8 A0A182RB65 A0A182TKG6 A0A182HQ94
PDB
5DTU
E-value=7.80069e-50,
Score=500
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
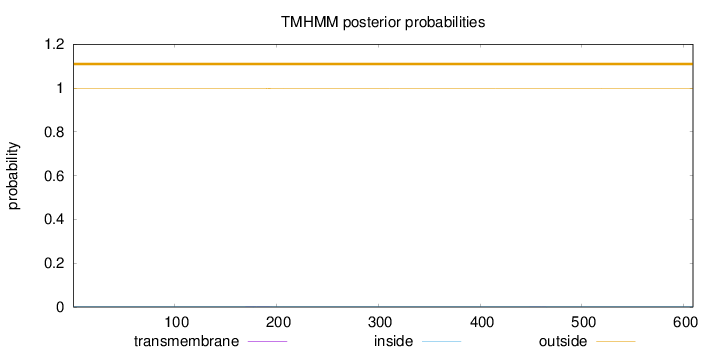
Length:
609
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05507
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00306
outside
1 - 609
Population Genetic Test Statistics
Pi
22.014702
Theta
20.629404
Tajima's D
0.285274
CLR
0.956964
CSRT
0.452027398630069
Interpretation
Uncertain
