Gene
KWMTBOMO04792 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009902
Annotation
partner_of_Y14_and_mago_[Bombyx_mori]
Full name
Partner of Y14 and mago
Alternative Name
Protein wibg homolog
Protein within the bgcn gene intron
Protein within the bgcn gene intron
Location in the cell
Nuclear Reliability : 3.363
Sequence
CDS
ATGTCAACACCAACGGCTTATGAGCACGACGCTGATGGGAGTAAATTCATTCCTGCAACTCAAAGACCTGATGGAACCTGGCGCAAGCCGAGAAGAATAAAAGAAGGTTATGTACCTCAAGAAGAGGTACCGTTGTATGAAAGCAAAGGAAAACAGTTCAGGGCCAGGCAAAATGATGGACTTCCGGTTGGACTTACACCTGAGATTGTAGCTCAAGCACAAAAGAAGAAAGGTCAACGAAGCACTATCCAACCTATACCTGGAATGATTATAACTGTTGAAAAGAAGAAAAAGAAAAAGAAGACAGTTACAGGTGTTGAAGAAGCTGCAGAAAAACTTGCCAAATGTGAAATACAAGAACCAACCGTACCTAGTCAGTCAGTACCGACTGAAAGTATATCTCAGTCTGATCCGACAAAACGCCTCAAGAATCTCAGAAAAAAGCTTAGAGAAATAGAGTTTCTAGAGGAAAAAATAAAAGCAGGGCTTCTTAAGAGCCCTGATAAAGATCAAAAAGAGAAAATGTCAAAAAAGAATGAAATTTTAAATGAAATTGACATTTTAGAGAATAGTATTCTATGA
Protein
MSTPTAYEHDADGSKFIPATQRPDGTWRKPRRIKEGYVPQEEVPLYESKGKQFRARQNDGLPVGLTPEIVAQAQKKKGQRSTIQPIPGMIITVEKKKKKKKTVTGVEEAAEKLAKCEIQEPTVPSQSVPTESISQSDPTKRLKNLRKKLREIEFLEEKIKAGLLKSPDKDQKEKMSKKNEILNEIDILENSIL
Summary
Description
Regulator of the exon junction complex (EJC), a multiprotein complex that associates immediately upstream of the exon-exon junction on mRNAs and serves as a positional landmarks for the intron exon structure of genes and directs post-transcriptional processes in the cytoplasm such as mRNA export, nonsense-mediated mRNA decay (NMD) or translation.
Regulator of the exon junction complex (EJC), a multiprotein complex that associates immediately upstream of the exon-exon junction on mRNAs and serves as a positional landmark for the intron exon structure of genes and directs post-transcriptional processes in the cytoplasm such as mRNA export, nonsense-mediated mRNA decay (NMD) or translation. Acts as an EJC disassembly factor by disrupting mature EJC from spliced mRNAs. Required for normal localization of osk mRNA to the posterior pole of the developing oocyte. Does not interact with the small ribosomal unit or components of the translation initiation complex. May not function in cap-dependent translation regulation.
Regulator of the exon junction complex (EJC), a multiprotein complex that associates immediately upstream of the exon-exon junction on mRNAs and serves as a positional landmark for the intron exon structure of genes and directs post-transcriptional processes in the cytoplasm such as mRNA export, nonsense-mediated mRNA decay (NMD) or translation. Acts as an EJC disassembly factor by disrupting mature EJC from spliced mRNAs. Required for normal localization of osk mRNA to the posterior pole of the developing oocyte. Does not interact with the small ribosomal unit or components of the translation initiation complex. May not function in cap-dependent translation regulation.
Subunit
Interacts (via N-terminus) with mago and tsu/Y14; the interaction is direct.
Interacts (via N-terminus) with mago and tsu/RBM8A; the interaction is direct.
Interacts (via N-terminus) with mago and tsu/RBM8A; the interaction is direct.
Similarity
Belongs to the pym family.
Belongs to the GroES chaperonin family.
Belongs to the GroES chaperonin family.
Keywords
Coiled coil
Complete proteome
Cytoplasm
Nucleus
Reference proteome
3D-structure
Feature
chain Partner of Y14 and mago
Uniprot
Q2F5J3
H9JK51
A0A2H1WK89
A0A194PQP3
A0A0N1I6C9
A0A1E1W9A3
+ More
A0A2A4JR05 S4PCU3 A0A212EV69 A0A0C9RUL8 A0A2J7PHK9 A0A1B6I007 A0A164Y948 A0A0P4XLG8 A0A0P5HXB9 A0A232EWZ9 A0A0P6BZD7 A0A0P5PAY5 K7J0X0 A0A0P5NYL1 A0A1B6FAA4 A0A0P6G7I6 A0A0Q9WPS1 A0A0P6GXI8 A0A1B0CB31 A0A1L8DCF5 A0A2A3E5I6 E9FYZ9 A0A088AA04 A0A0L7R6J4 A0A0Q5VW21 A0A0T6B3D3 P82804 A0A0K8TK69 B4NSP6 U5EZN3 A0A067QVG2 A0A0J9RL02 A0A1W4UMF6 E2C3R6 A0A1L8DC60 V5GKS4 D6WV11 A0A0R3NL25 A0A034WHC3 A0A0P4X209 A0A0R1DTI5 A0A1B6DNZ9 N6TPU1 A0A0P9AC03 A0A0K8UN40 T1PMB3 A0A1Y1N6T6 A0A1I8M3N1 A0A3L8E1R0 A0A2P8XEH2 A0A0M9ABK8 A0A026WEI4 A0A151WGC4 A0A158ND72 A0A195DJC2 E2AFB5 A0A151ILD8 A0A195FGS9 A0A2L2Y7J5 A0A1S3K099 A0A195B0S7 A0A2T7PAM5 A0A2R5LN60 A0A1W4XCU5
A0A2A4JR05 S4PCU3 A0A212EV69 A0A0C9RUL8 A0A2J7PHK9 A0A1B6I007 A0A164Y948 A0A0P4XLG8 A0A0P5HXB9 A0A232EWZ9 A0A0P6BZD7 A0A0P5PAY5 K7J0X0 A0A0P5NYL1 A0A1B6FAA4 A0A0P6G7I6 A0A0Q9WPS1 A0A0P6GXI8 A0A1B0CB31 A0A1L8DCF5 A0A2A3E5I6 E9FYZ9 A0A088AA04 A0A0L7R6J4 A0A0Q5VW21 A0A0T6B3D3 P82804 A0A0K8TK69 B4NSP6 U5EZN3 A0A067QVG2 A0A0J9RL02 A0A1W4UMF6 E2C3R6 A0A1L8DC60 V5GKS4 D6WV11 A0A0R3NL25 A0A034WHC3 A0A0P4X209 A0A0R1DTI5 A0A1B6DNZ9 N6TPU1 A0A0P9AC03 A0A0K8UN40 T1PMB3 A0A1Y1N6T6 A0A1I8M3N1 A0A3L8E1R0 A0A2P8XEH2 A0A0M9ABK8 A0A026WEI4 A0A151WGC4 A0A158ND72 A0A195DJC2 E2AFB5 A0A151ILD8 A0A195FGS9 A0A2L2Y7J5 A0A1S3K099 A0A195B0S7 A0A2T7PAM5 A0A2R5LN60 A0A1W4XCU5
Pubmed
EMBL
DQ311430
BABH01026667
ODYU01009206
SOQ53469.1
KQ459595
KPI95766.1
+ More
KQ461192 KPJ07059.1 GDQN01007464 JAT83590.1 NWSH01000843 PCG73883.1 GAIX01003971 JAA88589.1 AGBW02012233 OWR45379.1 GBYB01011290 GBYB01011291 JAG81057.1 JAG81058.1 NEVH01025135 PNF15815.1 GECU01027454 JAS80252.1 LRGB01000930 KZS15007.1 GDIP01240902 JAI82499.1 GDIQ01227407 JAK24318.1 NNAY01001806 OXU22870.1 GDIP01008340 JAM95375.1 GDIQ01137145 JAL14581.1 GDIQ01145946 JAL05780.1 GECZ01022600 JAS47169.1 GDIQ01040133 JAN54604.1 CH963850 KRF98138.1 GDIQ01027817 JAN66920.1 AJWK01004787 GFDF01009945 JAV04139.1 KZ288361 PBC26960.1 GL732527 EFX87683.1 KQ414646 KOC66444.1 CH954179 KQS63061.1 LJIG01016005 KRT81900.1 AF293388 AJ459405 AE013599 AY070957 AY071704 AAG00610.1 GDAI01002956 JAI14647.1 CH982397 GANO01000470 JAB59401.1 KK853329 KDR08434.1 CM002911 KMY96134.1 GL452364 EFN77302.1 GFDF01010119 JAV03965.1 GALX01006244 JAB62222.1 KQ971357 EFA08334.1 CM000071 KRT01572.1 GAKP01005397 JAC53555.1 GDRN01020005 JAI67817.1 CM000158 KRK00492.1 GEDC01009911 JAS27387.1 APGK01057053 KB741277 KB631617 ENN71265.1 ERL84715.1 CH902619 KPU75687.1 GDHF01024190 JAI28124.1 KA649048 AFP63677.1 GEZM01011168 JAV93614.1 QOIP01000001 RLU26393.1 PYGN01002487 PSN30407.1 KQ435700 KOX80496.1 KK107260 EZA54071.1 KQ983185 KYQ46855.1 ADTU01012469 KQ980800 KYN12917.1 GL439108 EFN67808.1 KQ977146 KYN05301.1 KQ981560 KYN39890.1 IAAA01018761 LAA04123.1 KQ976690 KYM78083.1 PZQS01000005 PVD30463.1 GGLE01006844 MBY10970.1
KQ461192 KPJ07059.1 GDQN01007464 JAT83590.1 NWSH01000843 PCG73883.1 GAIX01003971 JAA88589.1 AGBW02012233 OWR45379.1 GBYB01011290 GBYB01011291 JAG81057.1 JAG81058.1 NEVH01025135 PNF15815.1 GECU01027454 JAS80252.1 LRGB01000930 KZS15007.1 GDIP01240902 JAI82499.1 GDIQ01227407 JAK24318.1 NNAY01001806 OXU22870.1 GDIP01008340 JAM95375.1 GDIQ01137145 JAL14581.1 GDIQ01145946 JAL05780.1 GECZ01022600 JAS47169.1 GDIQ01040133 JAN54604.1 CH963850 KRF98138.1 GDIQ01027817 JAN66920.1 AJWK01004787 GFDF01009945 JAV04139.1 KZ288361 PBC26960.1 GL732527 EFX87683.1 KQ414646 KOC66444.1 CH954179 KQS63061.1 LJIG01016005 KRT81900.1 AF293388 AJ459405 AE013599 AY070957 AY071704 AAG00610.1 GDAI01002956 JAI14647.1 CH982397 GANO01000470 JAB59401.1 KK853329 KDR08434.1 CM002911 KMY96134.1 GL452364 EFN77302.1 GFDF01010119 JAV03965.1 GALX01006244 JAB62222.1 KQ971357 EFA08334.1 CM000071 KRT01572.1 GAKP01005397 JAC53555.1 GDRN01020005 JAI67817.1 CM000158 KRK00492.1 GEDC01009911 JAS27387.1 APGK01057053 KB741277 KB631617 ENN71265.1 ERL84715.1 CH902619 KPU75687.1 GDHF01024190 JAI28124.1 KA649048 AFP63677.1 GEZM01011168 JAV93614.1 QOIP01000001 RLU26393.1 PYGN01002487 PSN30407.1 KQ435700 KOX80496.1 KK107260 EZA54071.1 KQ983185 KYQ46855.1 ADTU01012469 KQ980800 KYN12917.1 GL439108 EFN67808.1 KQ977146 KYN05301.1 KQ981560 KYN39890.1 IAAA01018761 LAA04123.1 KQ976690 KYM78083.1 PZQS01000005 PVD30463.1 GGLE01006844 MBY10970.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000235965
+ More
UP000076858 UP000215335 UP000002358 UP000007798 UP000092461 UP000242457 UP000000305 UP000005203 UP000053825 UP000008711 UP000000803 UP000000304 UP000027135 UP000192221 UP000008237 UP000007266 UP000001819 UP000002282 UP000019118 UP000030742 UP000007801 UP000095301 UP000279307 UP000245037 UP000053105 UP000053097 UP000075809 UP000005205 UP000078492 UP000000311 UP000078542 UP000078541 UP000085678 UP000078540 UP000245119 UP000192223
UP000076858 UP000215335 UP000002358 UP000007798 UP000092461 UP000242457 UP000000305 UP000005203 UP000053825 UP000008711 UP000000803 UP000000304 UP000027135 UP000192221 UP000008237 UP000007266 UP000001819 UP000002282 UP000019118 UP000030742 UP000007801 UP000095301 UP000279307 UP000245037 UP000053105 UP000053097 UP000075809 UP000005205 UP000078492 UP000000311 UP000078542 UP000078541 UP000085678 UP000078540 UP000245119 UP000192223
Interpro
Gene 3D
CDD
ProteinModelPortal
Q2F5J3
H9JK51
A0A2H1WK89
A0A194PQP3
A0A0N1I6C9
A0A1E1W9A3
+ More
A0A2A4JR05 S4PCU3 A0A212EV69 A0A0C9RUL8 A0A2J7PHK9 A0A1B6I007 A0A164Y948 A0A0P4XLG8 A0A0P5HXB9 A0A232EWZ9 A0A0P6BZD7 A0A0P5PAY5 K7J0X0 A0A0P5NYL1 A0A1B6FAA4 A0A0P6G7I6 A0A0Q9WPS1 A0A0P6GXI8 A0A1B0CB31 A0A1L8DCF5 A0A2A3E5I6 E9FYZ9 A0A088AA04 A0A0L7R6J4 A0A0Q5VW21 A0A0T6B3D3 P82804 A0A0K8TK69 B4NSP6 U5EZN3 A0A067QVG2 A0A0J9RL02 A0A1W4UMF6 E2C3R6 A0A1L8DC60 V5GKS4 D6WV11 A0A0R3NL25 A0A034WHC3 A0A0P4X209 A0A0R1DTI5 A0A1B6DNZ9 N6TPU1 A0A0P9AC03 A0A0K8UN40 T1PMB3 A0A1Y1N6T6 A0A1I8M3N1 A0A3L8E1R0 A0A2P8XEH2 A0A0M9ABK8 A0A026WEI4 A0A151WGC4 A0A158ND72 A0A195DJC2 E2AFB5 A0A151ILD8 A0A195FGS9 A0A2L2Y7J5 A0A1S3K099 A0A195B0S7 A0A2T7PAM5 A0A2R5LN60 A0A1W4XCU5
A0A2A4JR05 S4PCU3 A0A212EV69 A0A0C9RUL8 A0A2J7PHK9 A0A1B6I007 A0A164Y948 A0A0P4XLG8 A0A0P5HXB9 A0A232EWZ9 A0A0P6BZD7 A0A0P5PAY5 K7J0X0 A0A0P5NYL1 A0A1B6FAA4 A0A0P6G7I6 A0A0Q9WPS1 A0A0P6GXI8 A0A1B0CB31 A0A1L8DCF5 A0A2A3E5I6 E9FYZ9 A0A088AA04 A0A0L7R6J4 A0A0Q5VW21 A0A0T6B3D3 P82804 A0A0K8TK69 B4NSP6 U5EZN3 A0A067QVG2 A0A0J9RL02 A0A1W4UMF6 E2C3R6 A0A1L8DC60 V5GKS4 D6WV11 A0A0R3NL25 A0A034WHC3 A0A0P4X209 A0A0R1DTI5 A0A1B6DNZ9 N6TPU1 A0A0P9AC03 A0A0K8UN40 T1PMB3 A0A1Y1N6T6 A0A1I8M3N1 A0A3L8E1R0 A0A2P8XEH2 A0A0M9ABK8 A0A026WEI4 A0A151WGC4 A0A158ND72 A0A195DJC2 E2AFB5 A0A151ILD8 A0A195FGS9 A0A2L2Y7J5 A0A1S3K099 A0A195B0S7 A0A2T7PAM5 A0A2R5LN60 A0A1W4XCU5
PDB
1RK8
E-value=7.2472e-14,
Score=183
Ontologies
PATHWAY
PANTHER
Topology
Subcellular location
Cytoplasm
Shuttles between the nucleus and the cytoplasm. Nuclear export is mediated by emb/Crm1 (By similarity). With evidence from 1 publications.
Nucleus Shuttles between the nucleus and the cytoplasm. Nuclear export is mediated by emb/Crm1 (By similarity). With evidence from 1 publications.
Nucleus Shuttles between the nucleus and the cytoplasm. Nuclear export is mediated by emb/Crm1 (By similarity). With evidence from 1 publications.
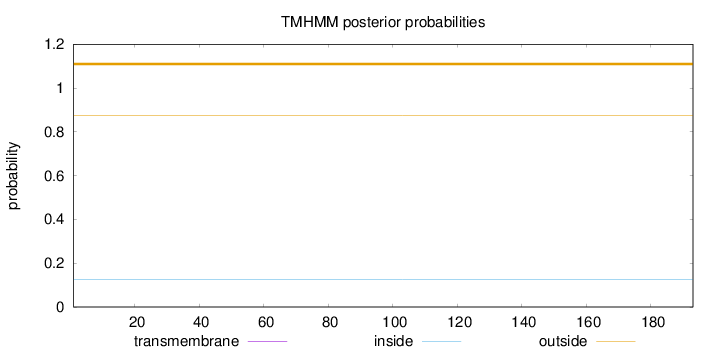
Length:
193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12557
outside
1 - 193
Population Genetic Test Statistics
Pi
26.387054
Theta
22.563663
Tajima's D
0.559719
CLR
2.667231
CSRT
0.539923003849808
Interpretation
Uncertain
