Gene
KWMTBOMO04789 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009841
Annotation
PREDICTED:_mitochondrial_matrix_protein_p33_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.099 Mitochondrial Reliability : 1.114
Sequence
CDS
ATGAGCGGCATAGTTAAAGCGACCCATAGAGTGCTGCAAACATGCGTTAAAACTGCGAACTTAAGCGGGTTGGTTAGTCGCACACAGACGGCTGTGAAACGCGACTTTACTAGGGGTATATGGCACATGAGCTGCTCTCGAAGGTTAGATGGGACCCTGGCTTCCACCAGCTTGCTACACAACCATTCCAATACTTGCAGCTGCGGCTGCGGGTTGAAGGCGCTTCACACGAAAGGTGAAAGAGAACTGGTAGAGTTCCTGACAGAAGAAATAGTTGCTGAACGCAAAGCACAGAAGGTCAAGTCTCTCCCTGCTGAAGTCGAAGGCTTTACTGTAAAGGGAGACGGTGCTGAAGTTGTTTTAACAAAGCAGCTTAAAGATGAAACAATTCGTGTCACATTCAACGTTAATCACACTGTGGACTCTGATGACTTTGAGGGTGATGTTCAGACTGAGAAGCAAGAGTTCTCTGAGATGCGTTCCAAACCTCAGTTTGAGGTTGACCTAGTCCGAGGAGACACCACCCTTGGTTTCACCTGCTCCTATCTTCAAGATCCACCTGCAGCTTCTGCTGATGAATATAACGATGTGTTCGGCATTGACGAAATAACCCTATATAAGGGTGAATGGAATGACAAAGTGTATGCAGTCGCTGGTGATGTGCTCGATGGGTATTTGTACGACCTACTCATGAACCTACTAGAAGAGAAGGGCATAAGCAATGAATTCGCGCAGAAGCTGTCGGACTTCAGTACTGCCTACGAGCACACGGCGTACATCAATCTTCTGGAGTCCATATCTAAATTCACCATCGGAAAATAA
Protein
MSGIVKATHRVLQTCVKTANLSGLVSRTQTAVKRDFTRGIWHMSCSRRLDGTLASTSLLHNHSNTCSCGCGLKALHTKGERELVEFLTEEIVAERKAQKVKSLPAEVEGFTVKGDGAEVVLTKQLKDETIRVTFNVNHTVDSDDFEGDVQTEKQEFSEMRSKPQFEVDLVRGDTTLGFTCSYLQDPPAASADEYNDVFGIDEITLYKGEWNDKVYAVAGDVLDGYLYDLLMNLLEEKGISNEFAQKLSDFSTAYEHTAYINLLESISKFTIGK
Summary
Uniprot
Q2F5P7
Q2F5P6
I4DJV4
A0A2A4JR39
I4DL90
A0A2H1VGS2
+ More
A0A194PS24 A0A0N1INI2 A0A212EV63 A0A0L0BQZ8 A0A0L7KW28 Q1HR26 A0A1S4FSN6 A0A0A1XRS1 A0A1B0D3F2 A0A1I8PT11 A0A034VZ40 A0A034VY66 A0A0K8TLA8 W8BPE6 A0A0K8U796 A0A1I8M7C2 A0A1A9ZAH6 T1PHT9 A0A1L8EDH7 A0A1A9YCY2 D3TLR5 A0A1B0BRW2 A0A1B0FHU4 A0A336M6W5 A0A336KYK4 B4KMZ8 T1D6F6 B0WPP0 A0A1A9UI95 B4MRZ7 B4LNA3 A0A1L8E2Y3 A0A067R160 R4UJ61 A0A0M4EW56 B4J787 B4P559 A0A1W4VCH4 A0APE6 A0A2J7R7J6 Q7JXC4 A0A0J9RF97 V5I9V3 A0APE4 B3MIE9 B4HMY6 A0APF0 D1GYM5 A0A0T6AYW1 D1GYM7 D1GYM3 D1GZ05 J3JXM0 D1GZ36 A0A1B6LSE6 A0A0M8ZVE5 N6T931 A0A1B6GSV1 A0A1B6IDI3 A0A2P8XJP2 A0A182F837 B3NML3 A0A0L7R1I1 T1DQC4 A0A2M4AJD6 A0A182QRU6 A0A1J1IM29 A0A2M4BWY1 A0A154PHD6 A0A2M4AH41 A0A2M4AHM2 W5J7Q5 A0A182VXN5 A0A2M3Z7I2 A0A1J1IQC7 A0A2M3Z7M2 A0A3B0J1H1 A0A2M3Z7W7 A0A182MDF8 A0A182N6G3 A0A088A0G9 A0A1Y9HEQ7 Q28Z43 B4GIA9 B4QBB9 A0A084W5B7 A0A1B6CV40 A0A182PF22 A0A182YIF2 A0A182XH52 A0A182KES2 A0A182I8A0 A0A182KMR6 D2CFW8
A0A194PS24 A0A0N1INI2 A0A212EV63 A0A0L0BQZ8 A0A0L7KW28 Q1HR26 A0A1S4FSN6 A0A0A1XRS1 A0A1B0D3F2 A0A1I8PT11 A0A034VZ40 A0A034VY66 A0A0K8TLA8 W8BPE6 A0A0K8U796 A0A1I8M7C2 A0A1A9ZAH6 T1PHT9 A0A1L8EDH7 A0A1A9YCY2 D3TLR5 A0A1B0BRW2 A0A1B0FHU4 A0A336M6W5 A0A336KYK4 B4KMZ8 T1D6F6 B0WPP0 A0A1A9UI95 B4MRZ7 B4LNA3 A0A1L8E2Y3 A0A067R160 R4UJ61 A0A0M4EW56 B4J787 B4P559 A0A1W4VCH4 A0APE6 A0A2J7R7J6 Q7JXC4 A0A0J9RF97 V5I9V3 A0APE4 B3MIE9 B4HMY6 A0APF0 D1GYM5 A0A0T6AYW1 D1GYM7 D1GYM3 D1GZ05 J3JXM0 D1GZ36 A0A1B6LSE6 A0A0M8ZVE5 N6T931 A0A1B6GSV1 A0A1B6IDI3 A0A2P8XJP2 A0A182F837 B3NML3 A0A0L7R1I1 T1DQC4 A0A2M4AJD6 A0A182QRU6 A0A1J1IM29 A0A2M4BWY1 A0A154PHD6 A0A2M4AH41 A0A2M4AHM2 W5J7Q5 A0A182VXN5 A0A2M3Z7I2 A0A1J1IQC7 A0A2M3Z7M2 A0A3B0J1H1 A0A2M3Z7W7 A0A182MDF8 A0A182N6G3 A0A088A0G9 A0A1Y9HEQ7 Q28Z43 B4GIA9 B4QBB9 A0A084W5B7 A0A1B6CV40 A0A182PF22 A0A182YIF2 A0A182XH52 A0A182KES2 A0A182I8A0 A0A182KMR6 D2CFW8
Pubmed
19121390
22651552
26354079
22118469
26108605
26227816
+ More
17204158 17510324 25830018 25348373 26369729 24495485 25315136 20353571 17994087 24330624 24845553 17550304 16951084 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19126864 26109357 26109356 22936249 18057021 22516182 23537049 29403074 20920257 23761445 15632085 24438588 25244985 20966253 18362917 19820115
17204158 17510324 25830018 25348373 26369729 24495485 25315136 20353571 17994087 24330624 24845553 17550304 16951084 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19126864 26109357 26109356 22936249 18057021 22516182 23537049 29403074 20920257 23761445 15632085 24438588 25244985 20966253 18362917 19820115
EMBL
DQ311376
ABD36320.1
BABH01026664
BABH01026665
BABH01026666
DQ311377
+ More
ABD36321.1 AK401572 BAM18194.1 NWSH01000843 PCG73880.1 AK402058 BAM18680.1 ODYU01002484 SOQ40026.1 KQ459595 KPI95763.1 KQ461192 KPJ07056.1 AGBW02012233 OWR45382.1 JRES01001589 KNC21664.1 JTDY01005143 KOB67330.1 DQ440268 CH477726 ABF18301.1 EAT36844.1 GBXI01000692 JAD13600.1 AJVK01023516 GAKP01012144 JAC46808.1 GAKP01012147 JAC46805.1 GDAI01002499 JAI15104.1 GAMC01003530 JAC03026.1 GDHF01029861 GDHF01013076 GDHF01008224 GDHF01005213 GDHF01003433 JAI22453.1 JAI39238.1 JAI44090.1 JAI47101.1 JAI48881.1 KA647690 AFP62319.1 GFDG01002019 JAV16780.1 EZ422367 ADD18643.1 JXJN01019356 CCAG010015795 UFQT01000643 SSX26042.1 UFQS01001157 UFQT01001157 SSX09288.1 SSX29190.1 CH933808 EDW09920.1 GALA01000078 JAA94774.1 DS232027 EDS32443.1 CH963850 EDW74886.1 CH940648 EDW61055.1 GFDF01000981 JAV13103.1 KK853008 KDR12558.1 KC632270 AGM32084.1 CP012524 ALC41970.1 CH916367 EDW02105.1 CM000158 EDW91760.1 AM294534 CAL26517.1 NEVH01006732 PNF36786.1 AE013599 AY094810 AM294527 AM294528 AM294529 AM294530 AM294531 AM294533 AM294535 AM294536 AM294537 FM245592 FM245593 FM245594 FM245595 FM245596 FM245597 FM245598 FM245599 FM245600 FM245601 AAF57817.1 AAM11163.1 CAL26506.1 CAL26510.1 CAL26511.1 CAL26512.1 CAL26514.1 CAL26516.1 CAL26518.1 CAL26519.1 CAL26520.1 CAR93518.1 CAR93519.1 CAR93520.1 CAR93521.1 CAR93522.1 CAR93523.1 CAR93524.1 CAR93525.1 CAR93526.1 CAR93527.1 CM002911 KMY94647.1 GALX01002285 JAB66181.1 AM294532 CAL26515.1 FN546610 FN546611 FN546613 FN546615 FN546617 FN546619 FN546620 CH902619 CBE66909.1 CBE66910.1 CBE66912.1 CBE66914.1 CBE66916.1 CBE66918.1 CBE66919.1 EDV36997.1 CH480816 EDW48336.1 AM294538 CAL26521.1 FN546614 CBE66913.1 LJIG01022490 KRT80300.1 FN546616 FN546618 CBE66915.1 CBE66917.1 FN546612 CBE66911.1 FN546744 CBE67043.1 BT127989 KB632230 AEE62951.1 ERL90385.1 FN546775 CBE67074.1 GEBQ01013355 JAT26622.1 KQ435863 KOX70403.1 APGK01039323 KB740969 ENN76744.1 GECZ01004346 JAS65423.1 GECU01022734 JAS84972.1 PYGN01001907 PSN32221.1 CH954179 EDV54952.1 KQ414667 KOC64698.1 GAMD01002614 JAA98976.1 GGFK01007563 MBW40884.1 AXCN02000286 CVRI01000055 CRL01303.1 GGFJ01008449 MBW57590.1 KQ434905 KZC11217.1 GGFK01006782 MBW40103.1 GGFK01006787 MBW40108.1 ADMH02002112 ETN58839.1 GGFM01003735 MBW24486.1 CRL01302.1 GGFM01003761 MBW24512.1 OUUW01000001 SPP75054.1 GGFM01003872 MBW24623.1 AXCM01004698 CM000071 EAL25772.1 CH479183 EDW36229.1 CM000362 EDX07543.1 ATLV01020573 KE525303 KFB45411.1 GEDC01021070 GEDC01019942 JAS16228.1 JAS17356.1 APCN01002684 DS497670 EFA13033.1
ABD36321.1 AK401572 BAM18194.1 NWSH01000843 PCG73880.1 AK402058 BAM18680.1 ODYU01002484 SOQ40026.1 KQ459595 KPI95763.1 KQ461192 KPJ07056.1 AGBW02012233 OWR45382.1 JRES01001589 KNC21664.1 JTDY01005143 KOB67330.1 DQ440268 CH477726 ABF18301.1 EAT36844.1 GBXI01000692 JAD13600.1 AJVK01023516 GAKP01012144 JAC46808.1 GAKP01012147 JAC46805.1 GDAI01002499 JAI15104.1 GAMC01003530 JAC03026.1 GDHF01029861 GDHF01013076 GDHF01008224 GDHF01005213 GDHF01003433 JAI22453.1 JAI39238.1 JAI44090.1 JAI47101.1 JAI48881.1 KA647690 AFP62319.1 GFDG01002019 JAV16780.1 EZ422367 ADD18643.1 JXJN01019356 CCAG010015795 UFQT01000643 SSX26042.1 UFQS01001157 UFQT01001157 SSX09288.1 SSX29190.1 CH933808 EDW09920.1 GALA01000078 JAA94774.1 DS232027 EDS32443.1 CH963850 EDW74886.1 CH940648 EDW61055.1 GFDF01000981 JAV13103.1 KK853008 KDR12558.1 KC632270 AGM32084.1 CP012524 ALC41970.1 CH916367 EDW02105.1 CM000158 EDW91760.1 AM294534 CAL26517.1 NEVH01006732 PNF36786.1 AE013599 AY094810 AM294527 AM294528 AM294529 AM294530 AM294531 AM294533 AM294535 AM294536 AM294537 FM245592 FM245593 FM245594 FM245595 FM245596 FM245597 FM245598 FM245599 FM245600 FM245601 AAF57817.1 AAM11163.1 CAL26506.1 CAL26510.1 CAL26511.1 CAL26512.1 CAL26514.1 CAL26516.1 CAL26518.1 CAL26519.1 CAL26520.1 CAR93518.1 CAR93519.1 CAR93520.1 CAR93521.1 CAR93522.1 CAR93523.1 CAR93524.1 CAR93525.1 CAR93526.1 CAR93527.1 CM002911 KMY94647.1 GALX01002285 JAB66181.1 AM294532 CAL26515.1 FN546610 FN546611 FN546613 FN546615 FN546617 FN546619 FN546620 CH902619 CBE66909.1 CBE66910.1 CBE66912.1 CBE66914.1 CBE66916.1 CBE66918.1 CBE66919.1 EDV36997.1 CH480816 EDW48336.1 AM294538 CAL26521.1 FN546614 CBE66913.1 LJIG01022490 KRT80300.1 FN546616 FN546618 CBE66915.1 CBE66917.1 FN546612 CBE66911.1 FN546744 CBE67043.1 BT127989 KB632230 AEE62951.1 ERL90385.1 FN546775 CBE67074.1 GEBQ01013355 JAT26622.1 KQ435863 KOX70403.1 APGK01039323 KB740969 ENN76744.1 GECZ01004346 JAS65423.1 GECU01022734 JAS84972.1 PYGN01001907 PSN32221.1 CH954179 EDV54952.1 KQ414667 KOC64698.1 GAMD01002614 JAA98976.1 GGFK01007563 MBW40884.1 AXCN02000286 CVRI01000055 CRL01303.1 GGFJ01008449 MBW57590.1 KQ434905 KZC11217.1 GGFK01006782 MBW40103.1 GGFK01006787 MBW40108.1 ADMH02002112 ETN58839.1 GGFM01003735 MBW24486.1 CRL01302.1 GGFM01003761 MBW24512.1 OUUW01000001 SPP75054.1 GGFM01003872 MBW24623.1 AXCM01004698 CM000071 EAL25772.1 CH479183 EDW36229.1 CM000362 EDX07543.1 ATLV01020573 KE525303 KFB45411.1 GEDC01021070 GEDC01019942 JAS16228.1 JAS17356.1 APCN01002684 DS497670 EFA13033.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037069
+ More
UP000037510 UP000008820 UP000092462 UP000095300 UP000095301 UP000092445 UP000092443 UP000092460 UP000092444 UP000009192 UP000002320 UP000078200 UP000007798 UP000008792 UP000027135 UP000092553 UP000001070 UP000002282 UP000192221 UP000235965 UP000000803 UP000007801 UP000001292 UP000030742 UP000053105 UP000019118 UP000245037 UP000069272 UP000008711 UP000053825 UP000075886 UP000183832 UP000076502 UP000000673 UP000075920 UP000268350 UP000075883 UP000075884 UP000005203 UP000075900 UP000001819 UP000008744 UP000000304 UP000030765 UP000075885 UP000076408 UP000076407 UP000075881 UP000075840 UP000075882 UP000007266
UP000037510 UP000008820 UP000092462 UP000095300 UP000095301 UP000092445 UP000092443 UP000092460 UP000092444 UP000009192 UP000002320 UP000078200 UP000007798 UP000008792 UP000027135 UP000092553 UP000001070 UP000002282 UP000192221 UP000235965 UP000000803 UP000007801 UP000001292 UP000030742 UP000053105 UP000019118 UP000245037 UP000069272 UP000008711 UP000053825 UP000075886 UP000183832 UP000076502 UP000000673 UP000075920 UP000268350 UP000075883 UP000075884 UP000005203 UP000075900 UP000001819 UP000008744 UP000000304 UP000030765 UP000075885 UP000076408 UP000076407 UP000075881 UP000075840 UP000075882 UP000007266
Interpro
Gene 3D
CDD
ProteinModelPortal
Q2F5P7
Q2F5P6
I4DJV4
A0A2A4JR39
I4DL90
A0A2H1VGS2
+ More
A0A194PS24 A0A0N1INI2 A0A212EV63 A0A0L0BQZ8 A0A0L7KW28 Q1HR26 A0A1S4FSN6 A0A0A1XRS1 A0A1B0D3F2 A0A1I8PT11 A0A034VZ40 A0A034VY66 A0A0K8TLA8 W8BPE6 A0A0K8U796 A0A1I8M7C2 A0A1A9ZAH6 T1PHT9 A0A1L8EDH7 A0A1A9YCY2 D3TLR5 A0A1B0BRW2 A0A1B0FHU4 A0A336M6W5 A0A336KYK4 B4KMZ8 T1D6F6 B0WPP0 A0A1A9UI95 B4MRZ7 B4LNA3 A0A1L8E2Y3 A0A067R160 R4UJ61 A0A0M4EW56 B4J787 B4P559 A0A1W4VCH4 A0APE6 A0A2J7R7J6 Q7JXC4 A0A0J9RF97 V5I9V3 A0APE4 B3MIE9 B4HMY6 A0APF0 D1GYM5 A0A0T6AYW1 D1GYM7 D1GYM3 D1GZ05 J3JXM0 D1GZ36 A0A1B6LSE6 A0A0M8ZVE5 N6T931 A0A1B6GSV1 A0A1B6IDI3 A0A2P8XJP2 A0A182F837 B3NML3 A0A0L7R1I1 T1DQC4 A0A2M4AJD6 A0A182QRU6 A0A1J1IM29 A0A2M4BWY1 A0A154PHD6 A0A2M4AH41 A0A2M4AHM2 W5J7Q5 A0A182VXN5 A0A2M3Z7I2 A0A1J1IQC7 A0A2M3Z7M2 A0A3B0J1H1 A0A2M3Z7W7 A0A182MDF8 A0A182N6G3 A0A088A0G9 A0A1Y9HEQ7 Q28Z43 B4GIA9 B4QBB9 A0A084W5B7 A0A1B6CV40 A0A182PF22 A0A182YIF2 A0A182XH52 A0A182KES2 A0A182I8A0 A0A182KMR6 D2CFW8
A0A194PS24 A0A0N1INI2 A0A212EV63 A0A0L0BQZ8 A0A0L7KW28 Q1HR26 A0A1S4FSN6 A0A0A1XRS1 A0A1B0D3F2 A0A1I8PT11 A0A034VZ40 A0A034VY66 A0A0K8TLA8 W8BPE6 A0A0K8U796 A0A1I8M7C2 A0A1A9ZAH6 T1PHT9 A0A1L8EDH7 A0A1A9YCY2 D3TLR5 A0A1B0BRW2 A0A1B0FHU4 A0A336M6W5 A0A336KYK4 B4KMZ8 T1D6F6 B0WPP0 A0A1A9UI95 B4MRZ7 B4LNA3 A0A1L8E2Y3 A0A067R160 R4UJ61 A0A0M4EW56 B4J787 B4P559 A0A1W4VCH4 A0APE6 A0A2J7R7J6 Q7JXC4 A0A0J9RF97 V5I9V3 A0APE4 B3MIE9 B4HMY6 A0APF0 D1GYM5 A0A0T6AYW1 D1GYM7 D1GYM3 D1GZ05 J3JXM0 D1GZ36 A0A1B6LSE6 A0A0M8ZVE5 N6T931 A0A1B6GSV1 A0A1B6IDI3 A0A2P8XJP2 A0A182F837 B3NML3 A0A0L7R1I1 T1DQC4 A0A2M4AJD6 A0A182QRU6 A0A1J1IM29 A0A2M4BWY1 A0A154PHD6 A0A2M4AH41 A0A2M4AHM2 W5J7Q5 A0A182VXN5 A0A2M3Z7I2 A0A1J1IQC7 A0A2M3Z7M2 A0A3B0J1H1 A0A2M3Z7W7 A0A182MDF8 A0A182N6G3 A0A088A0G9 A0A1Y9HEQ7 Q28Z43 B4GIA9 B4QBB9 A0A084W5B7 A0A1B6CV40 A0A182PF22 A0A182YIF2 A0A182XH52 A0A182KES2 A0A182I8A0 A0A182KMR6 D2CFW8
PDB
1P32
E-value=1.46977e-23,
Score=269
Ontologies
GO
Topology
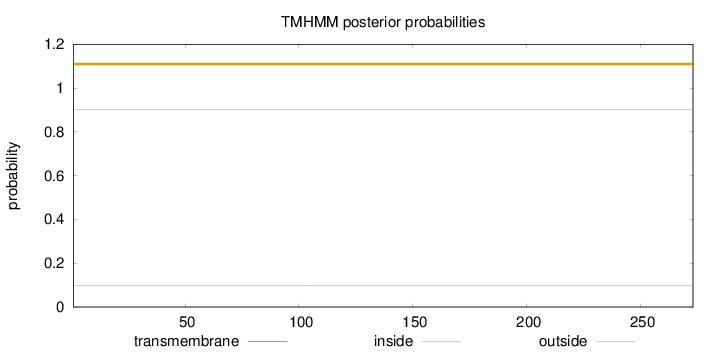
Length:
273
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00316
Exp number, first 60 AAs:
0.00079
Total prob of N-in:
0.09869
outside
1 - 273
Population Genetic Test Statistics
Pi
22.60431
Theta
18.145384
Tajima's D
-1.140092
CLR
1.141082
CSRT
0.111344432778361
Interpretation
Uncertain
