Gene
KWMTBOMO04788
Pre Gene Modal
BGIBMGA009844
Annotation
putative_amyloid_beta_precursor_protein_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.006 Nuclear Reliability : 1.089
Sequence
CDS
ATGCTGCAGGAGAAGAAGTTGTGTATTTTACACACGGAGCTATCAGAGTTGGATGTCTTCGAGAGGCTGTTGCGGTTCGCAGGTTGCCAGCTGAAACTCCTCGAGTGCTTCCAGGCAGTCATAGAACACTGTTCGAAGCTGTCAACCGAACTAGCGACCGCTTACGTTGAGAAGTGCAATGCGAACGTCAGATCTAGGAACTCTCTGATACAGCTCGGGTTGAGGCTCGGTGGATTCTTGAACGAAGCCGGGTGGTACGCGGACGCGCAGACCGTCCTCCTCAAGTGCAGGAGCCTCTGTCAGGCACAGCCGCAGTCGACGTACTACAAAAGACTGACCCTCGAATGTTGTCATAGATTATTGAACACACAATCAGCGTATTGCTGTTTCCCCGCCGCGGCCGAGACGTACTCGCTGGCCTTAAAGCTATTGGGCATCACTGAAGACGTCGCCGAGCTTCCCGAATGCTCATTCAGTGTGGCGGAAACTTGCGACGTAGCCTCGCAGACTGACGGCGACATCATGAATCCATGCGAGAGGGAGGGGGAGATATCGGAGGAGTGCTGCAGATGCGGCAGCACCGGCACGTGGGGCTCCAGCAGTCTGCTGGCGCGGCTGTGCAAGGAGTTCAGCTCGCTGTTCTTCGTCCGCTGCGACTACGGAGCCGCCTACACGTGGAGCATGAGGGCGCTCAAGCTCCTCACTCCGCACACGCCCGTCAAGATCACGATCGAGGTGCTGCGCGCGTGCGGCAAGGCGTGCGTGGTGAAGCGGCGGTTCCGCGGCGCCGGCCTGCTCGTGCGCCACGCCGCCGCGCTCGCCCGCGCCGCCTACGGGCCGCGCCACCCGCGACACGCCGCCGCGCTGCTCGACCTGGGCTTCTACCTGCTCAACGTCGACTGCGTGCTGCACAGCGTCGCCGTGTACGAGGAGGCGCTGGCCGCGCTGAGCGCCGCGTTCGGCCGCCACAGTCTGCACGTGGCCACCGCGCAGGAGGACCTCGCCTACGCGCTCTACGTGCTGGAGTACTCCTCGGGACGCTTCTACAAGGCGAGGGAGCACGCCGAGAGGGCCATTAGGATCATACAGAACCTGGTGCCCCCTGACCACTTGCTCCTGGCGTCAGCGAAACGCGTCAAGGCTCTCATACTGGAGGAGATAGCCCTGGACACGCCCGCGCGCCAGGACATGAGCGAGCCGAGTCTGCTGGAGGAGTCGGAGGCGCTGCATAAGGCCGCGCTGGAGCTGTCCATGCTGGCGTTCGGCGAGAAGAACGTGCAGACCGCGAAGCACTACGGGAACCTGGGCCGACTCTACCAGAGCATGCACAAGTTCCACGACGCGGAGACGATGCACCTGAAGGCGATAGCGATAAAGGAGGAGCTGCTGGGCCCCGAGGACTACGAGGTGGGGCTCAGCATCGGCCACCTCGCCTCGCTCTACAACTACCACATGAACATGCACCGCGCCGCAGAGAAGCTCTACCTGCGCTCCATACACATAAGTGTGAAGTTGTTCGGCGAGCGCTACTCGGGGCTGGAGTACGACTACCGCGGGCTGGTGCACGTCTGCACGCAGCTCAAGCGGCACGACGCCGCCGCACACTACGCGCGCGTGCTGCAGCGCTGGCAACAACTTCGTGAGAAATCAAAAGCGGAGCAACAGCCGGCCCTCGGCGACGAGGAAGTGCTGTCGTTAGAAGAACTACAAGCGAAACTGTTCGCGCCCAACGACTGA
Protein
MLQEKKLCILHTELSELDVFERLLRFAGCQLKLLECFQAVIEHCSKLSTELATAYVEKCNANVRSRNSLIQLGLRLGGFLNEAGWYADAQTVLLKCRSLCQAQPQSTYYKRLTLECCHRLLNTQSAYCCFPAAAETYSLALKLLGITEDVAELPECSFSVAETCDVASQTDGDIMNPCEREGEISEECCRCGSTGTWGSSSLLARLCKEFSSLFFVRCDYGAAYTWSMRALKLLTPHTPVKITIEVLRACGKACVVKRRFRGAGLLVRHAAALARAAYGPRHPRHAAALLDLGFYLLNVDCVLHSVAVYEEALAALSAAFGRHSLHVATAQEDLAYALYVLEYSSGRFYKAREHAERAIRIIQNLVPPDHLLLASAKRVKALILEEIALDTPARQDMSEPSLLEESEALHKAALELSMLAFGEKNVQTAKHYGNLGRLYQSMHKFHDAETMHLKAIAIKEELLGPEDYEVGLSIGHLASLYNYHMNMHRAAEKLYLRSIHISVKLFGERYSGLEYDYRGLVHVCTQLKRHDAAAHYARVLQRWQQLREKSKAEQQPALGDEEVLSLEELQAKLFAPND
Summary
Uniprot
H9JJZ3
A0A212EVA5
A0A194PR25
A0A2H1V1A0
A0A2A4JIE5
A0A0L7L717
+ More
A0A2J7QZD6 E2C9Z9 A0A026WB75 A0A067QHR8 E2AJJ8 A0A195D495 A0A232EF84 A0A151XFH9 A0A195E290 F4WVQ5 A0A195AUS7 A0A158NHI5 A0A1B6GNC6 A0A195FWU6 A0A0C9PH99 A0A069DVL2 A0A224X9Z5 A0A023EYX0 T1HTM5 E9I864 A0A088A234 A0A2A3ELI0 A0A1Y1NJ10 A0A1Y1NHY2 D6WXC0 A0A293M340 A0A2R5LL80 V5HSC7 A0A0A9Y3S2 T1JC70 A0A154PRF3 A0A131XIK6 A0A286X8N7 A0A1E1X5Z3 H3BB79 A0A0M8ZWC0 V9KCF8 L7M7J8 A0A2Y9DNE9 A0A099Z9C1 A0A0D9R101 A0A1L8HG21 A0A3P4S593 R0LXC7 A0A131YHC0 A0A091VPC2 A0A091KHZ1 A0A091W586 A0A091PMT6 A0A091RXK9 L8IUK2 E1B7N1 A0A091M183 A0A087VKA6 A0A093C5F8 A0A0A0AUC3 A0A091NTQ8 A0A091T993 A0A093NGY0 A0A094KJF8 A0A091PDJ5 G1SX62 A0A093C4S7 A0A091L6Z4 A0A091IWH3 A0A091QFX1 G3TI80 A0A1W4X0A5 I3MH61 F7HXY0 A0A093I514 U3DI47 A0A093I891 A0A384CGX9 A0A091U237 G3VE34 A0A1S3EZH0 A0A250Y397 A0A2K6BU20 A0A2K6JNC3 K9J203 G1QHR1 A0A2K5ZPZ1 F1S207 F6XXW5 A0A2R9ARL1 A0A096P4Z6 A0A2K6Q688 H0XRA2 A0A1U7SSE5
A0A2J7QZD6 E2C9Z9 A0A026WB75 A0A067QHR8 E2AJJ8 A0A195D495 A0A232EF84 A0A151XFH9 A0A195E290 F4WVQ5 A0A195AUS7 A0A158NHI5 A0A1B6GNC6 A0A195FWU6 A0A0C9PH99 A0A069DVL2 A0A224X9Z5 A0A023EYX0 T1HTM5 E9I864 A0A088A234 A0A2A3ELI0 A0A1Y1NJ10 A0A1Y1NHY2 D6WXC0 A0A293M340 A0A2R5LL80 V5HSC7 A0A0A9Y3S2 T1JC70 A0A154PRF3 A0A131XIK6 A0A286X8N7 A0A1E1X5Z3 H3BB79 A0A0M8ZWC0 V9KCF8 L7M7J8 A0A2Y9DNE9 A0A099Z9C1 A0A0D9R101 A0A1L8HG21 A0A3P4S593 R0LXC7 A0A131YHC0 A0A091VPC2 A0A091KHZ1 A0A091W586 A0A091PMT6 A0A091RXK9 L8IUK2 E1B7N1 A0A091M183 A0A087VKA6 A0A093C5F8 A0A0A0AUC3 A0A091NTQ8 A0A091T993 A0A093NGY0 A0A094KJF8 A0A091PDJ5 G1SX62 A0A093C4S7 A0A091L6Z4 A0A091IWH3 A0A091QFX1 G3TI80 A0A1W4X0A5 I3MH61 F7HXY0 A0A093I514 U3DI47 A0A093I891 A0A384CGX9 A0A091U237 G3VE34 A0A1S3EZH0 A0A250Y397 A0A2K6BU20 A0A2K6JNC3 K9J203 G1QHR1 A0A2K5ZPZ1 F1S207 F6XXW5 A0A2R9ARL1 A0A096P4Z6 A0A2K6Q688 H0XRA2 A0A1U7SSE5
Pubmed
19121390
22118469
26354079
26227816
20798317
24508170
+ More
30249741 24845553 28648823 21719571 21347285 26334808 25474469 21282665 28004739 18362917 19820115 25765539 25401762 26823975 28049606 21993624 28503490 9215903 24402279 25576852 27762356 26830274 22751099 19393038 25243066 21709235 28087693 30723633 19892987 22722832 25362486
30249741 24845553 28648823 21719571 21347285 26334808 25474469 21282665 28004739 18362917 19820115 25765539 25401762 26823975 28049606 21993624 28503490 9215903 24402279 25576852 27762356 26830274 22751099 19393038 25243066 21709235 28087693 30723633 19892987 22722832 25362486
EMBL
BABH01026658
BABH01026659
AGBW02012233
OWR45384.1
KQ459595
KPI95762.1
+ More
ODYU01000217 SOQ34617.1 NWSH01001449 PCG71182.1 JTDY01002596 KOB71139.1 NEVH01009069 PNF33960.1 GL453916 EFN75236.1 KK107293 QOIP01000005 EZA53347.1 RLU22376.1 KK853603 KDR06362.1 GL440027 EFN66393.1 KQ976870 KYN07725.1 NNAY01005088 OXU17020.1 KQ982194 KYQ59040.1 KQ979763 KYN19258.1 GL888393 EGI61725.1 KQ976737 KYM75931.1 ADTU01015833 ADTU01015834 GECZ01005821 JAS63948.1 KQ981208 KYN44901.1 GBYB01000228 JAG69995.1 GBGD01001048 JAC87841.1 GFTR01007239 JAW09187.1 GBBI01004309 JAC14403.1 ACPB03015394 GL761471 EFZ23235.1 KZ288215 PBC32560.1 GEZM01006551 JAV95727.1 GEZM01006552 JAV95726.1 KQ971361 EFA08821.1 GFWV01010603 MAA35332.1 GGLE01006072 MBY10198.1 GANP01007470 JAB76998.1 GBHO01029961 GBHO01025902 GBHO01025896 GBHO01025890 GBHO01019457 GBHO01019454 GBHO01019451 GBHO01019450 GBHO01016121 GDHC01006670 GDHC01005876 GDHC01004498 JAG13643.1 JAG17702.1 JAG17708.1 JAG17714.1 JAG24147.1 JAG24150.1 JAG24153.1 JAG24154.1 JAG27483.1 JAQ11959.1 JAQ12753.1 JAQ14131.1 JH432062 KQ435078 KZC14496.1 GEFH01002234 JAP66347.1 AAKN02033077 AAKN02033078 AAKN02033079 GFAC01004499 JAT94689.1 AFYH01004275 KQ435821 KOX72445.1 JW863067 AFO95584.1 GACK01005009 JAA60025.1 KL891046 KGL78356.1 AQIB01117157 AQIB01117158 AQIB01117159 CM004468 OCT94981.1 CYRY02046611 VCX42367.1 KB742712 EOB05153.1 GEDV01010655 JAP77902.1 KL409726 KFQ91523.1 KK742173 KFP39108.1 KK734761 KFR10639.1 KK675633 KFQ08616.1 KK935449 KFQ46939.1 JH880797 ELR58847.1 KK518431 KFP65583.1 KL496500 KFO13048.1 KL237229 KFV09638.1 KL873333 KGL98184.1 KL374526 KFP82845.1 KK441628 KFQ70362.1 KL224565 KFW61255.1 KL350735 KFZ58735.1 KK657124 KFQ05675.1 AAGW02039248 AAGW02039249 KL456318 KFV09443.1 KL303123 KFP51627.1 KK501002 KFP11903.1 KK698615 KFQ25907.1 AGTP01022487 AGTP01022488 AGTP01022489 GAMT01007321 GAMS01001727 GAMR01001008 GAMP01003854 GAMP01003853 JAB04540.1 JAB21409.1 JAB32924.1 JAB48901.1 KL214799 KFV61821.1 GAMQ01003498 JAB38353.1 KK589023 KFV98919.1 KK495740 KFQ65472.1 AEFK01160814 AEFK01160815 GFFV01001910 JAV38035.1 GABZ01005186 JAA48339.1 ADFV01107551 ADFV01107552 ADFV01107553 ADFV01107554 AEMK02000080 DQIR01197059 HDB52536.1 AJFE02046695 AJFE02046696 AJFE02046697 AJFE02046698 AJFE02046699 AJFE02046700 AHZZ02010261 AAQR03121782 AAQR03121783 AAQR03121784 AAQR03121785 AAQR03121786 AAQR03121787 AAQR03121788 AAQR03121789 AAQR03121790 AAQR03121791
ODYU01000217 SOQ34617.1 NWSH01001449 PCG71182.1 JTDY01002596 KOB71139.1 NEVH01009069 PNF33960.1 GL453916 EFN75236.1 KK107293 QOIP01000005 EZA53347.1 RLU22376.1 KK853603 KDR06362.1 GL440027 EFN66393.1 KQ976870 KYN07725.1 NNAY01005088 OXU17020.1 KQ982194 KYQ59040.1 KQ979763 KYN19258.1 GL888393 EGI61725.1 KQ976737 KYM75931.1 ADTU01015833 ADTU01015834 GECZ01005821 JAS63948.1 KQ981208 KYN44901.1 GBYB01000228 JAG69995.1 GBGD01001048 JAC87841.1 GFTR01007239 JAW09187.1 GBBI01004309 JAC14403.1 ACPB03015394 GL761471 EFZ23235.1 KZ288215 PBC32560.1 GEZM01006551 JAV95727.1 GEZM01006552 JAV95726.1 KQ971361 EFA08821.1 GFWV01010603 MAA35332.1 GGLE01006072 MBY10198.1 GANP01007470 JAB76998.1 GBHO01029961 GBHO01025902 GBHO01025896 GBHO01025890 GBHO01019457 GBHO01019454 GBHO01019451 GBHO01019450 GBHO01016121 GDHC01006670 GDHC01005876 GDHC01004498 JAG13643.1 JAG17702.1 JAG17708.1 JAG17714.1 JAG24147.1 JAG24150.1 JAG24153.1 JAG24154.1 JAG27483.1 JAQ11959.1 JAQ12753.1 JAQ14131.1 JH432062 KQ435078 KZC14496.1 GEFH01002234 JAP66347.1 AAKN02033077 AAKN02033078 AAKN02033079 GFAC01004499 JAT94689.1 AFYH01004275 KQ435821 KOX72445.1 JW863067 AFO95584.1 GACK01005009 JAA60025.1 KL891046 KGL78356.1 AQIB01117157 AQIB01117158 AQIB01117159 CM004468 OCT94981.1 CYRY02046611 VCX42367.1 KB742712 EOB05153.1 GEDV01010655 JAP77902.1 KL409726 KFQ91523.1 KK742173 KFP39108.1 KK734761 KFR10639.1 KK675633 KFQ08616.1 KK935449 KFQ46939.1 JH880797 ELR58847.1 KK518431 KFP65583.1 KL496500 KFO13048.1 KL237229 KFV09638.1 KL873333 KGL98184.1 KL374526 KFP82845.1 KK441628 KFQ70362.1 KL224565 KFW61255.1 KL350735 KFZ58735.1 KK657124 KFQ05675.1 AAGW02039248 AAGW02039249 KL456318 KFV09443.1 KL303123 KFP51627.1 KK501002 KFP11903.1 KK698615 KFQ25907.1 AGTP01022487 AGTP01022488 AGTP01022489 GAMT01007321 GAMS01001727 GAMR01001008 GAMP01003854 GAMP01003853 JAB04540.1 JAB21409.1 JAB32924.1 JAB48901.1 KL214799 KFV61821.1 GAMQ01003498 JAB38353.1 KK589023 KFV98919.1 KK495740 KFQ65472.1 AEFK01160814 AEFK01160815 GFFV01001910 JAV38035.1 GABZ01005186 JAA48339.1 ADFV01107551 ADFV01107552 ADFV01107553 ADFV01107554 AEMK02000080 DQIR01197059 HDB52536.1 AJFE02046695 AJFE02046696 AJFE02046697 AJFE02046698 AJFE02046699 AJFE02046700 AHZZ02010261 AAQR03121782 AAQR03121783 AAQR03121784 AAQR03121785 AAQR03121786 AAQR03121787 AAQR03121788 AAQR03121789 AAQR03121790 AAQR03121791
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000037510
UP000235965
+ More
UP000008237 UP000053097 UP000279307 UP000027135 UP000000311 UP000078542 UP000215335 UP000075809 UP000078492 UP000007755 UP000078540 UP000005205 UP000078541 UP000015103 UP000005203 UP000242457 UP000007266 UP000076502 UP000005447 UP000008672 UP000053105 UP000248480 UP000053641 UP000029965 UP000186698 UP000053283 UP000053605 UP000009136 UP000053858 UP000054081 UP000001811 UP000053119 UP000007646 UP000192223 UP000005215 UP000008225 UP000053875 UP000261680 UP000007648 UP000081671 UP000233120 UP000233180 UP000001073 UP000233140 UP000008227 UP000002281 UP000240080 UP000028761 UP000233200 UP000005225 UP000189704
UP000008237 UP000053097 UP000279307 UP000027135 UP000000311 UP000078542 UP000215335 UP000075809 UP000078492 UP000007755 UP000078540 UP000005205 UP000078541 UP000015103 UP000005203 UP000242457 UP000007266 UP000076502 UP000005447 UP000008672 UP000053105 UP000248480 UP000053641 UP000029965 UP000186698 UP000053283 UP000053605 UP000009136 UP000053858 UP000054081 UP000001811 UP000053119 UP000007646 UP000192223 UP000005215 UP000008225 UP000053875 UP000261680 UP000007648 UP000081671 UP000233120 UP000233180 UP000001073 UP000233140 UP000008227 UP000002281 UP000240080 UP000028761 UP000233200 UP000005225 UP000189704
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9JJZ3
A0A212EVA5
A0A194PR25
A0A2H1V1A0
A0A2A4JIE5
A0A0L7L717
+ More
A0A2J7QZD6 E2C9Z9 A0A026WB75 A0A067QHR8 E2AJJ8 A0A195D495 A0A232EF84 A0A151XFH9 A0A195E290 F4WVQ5 A0A195AUS7 A0A158NHI5 A0A1B6GNC6 A0A195FWU6 A0A0C9PH99 A0A069DVL2 A0A224X9Z5 A0A023EYX0 T1HTM5 E9I864 A0A088A234 A0A2A3ELI0 A0A1Y1NJ10 A0A1Y1NHY2 D6WXC0 A0A293M340 A0A2R5LL80 V5HSC7 A0A0A9Y3S2 T1JC70 A0A154PRF3 A0A131XIK6 A0A286X8N7 A0A1E1X5Z3 H3BB79 A0A0M8ZWC0 V9KCF8 L7M7J8 A0A2Y9DNE9 A0A099Z9C1 A0A0D9R101 A0A1L8HG21 A0A3P4S593 R0LXC7 A0A131YHC0 A0A091VPC2 A0A091KHZ1 A0A091W586 A0A091PMT6 A0A091RXK9 L8IUK2 E1B7N1 A0A091M183 A0A087VKA6 A0A093C5F8 A0A0A0AUC3 A0A091NTQ8 A0A091T993 A0A093NGY0 A0A094KJF8 A0A091PDJ5 G1SX62 A0A093C4S7 A0A091L6Z4 A0A091IWH3 A0A091QFX1 G3TI80 A0A1W4X0A5 I3MH61 F7HXY0 A0A093I514 U3DI47 A0A093I891 A0A384CGX9 A0A091U237 G3VE34 A0A1S3EZH0 A0A250Y397 A0A2K6BU20 A0A2K6JNC3 K9J203 G1QHR1 A0A2K5ZPZ1 F1S207 F6XXW5 A0A2R9ARL1 A0A096P4Z6 A0A2K6Q688 H0XRA2 A0A1U7SSE5
A0A2J7QZD6 E2C9Z9 A0A026WB75 A0A067QHR8 E2AJJ8 A0A195D495 A0A232EF84 A0A151XFH9 A0A195E290 F4WVQ5 A0A195AUS7 A0A158NHI5 A0A1B6GNC6 A0A195FWU6 A0A0C9PH99 A0A069DVL2 A0A224X9Z5 A0A023EYX0 T1HTM5 E9I864 A0A088A234 A0A2A3ELI0 A0A1Y1NJ10 A0A1Y1NHY2 D6WXC0 A0A293M340 A0A2R5LL80 V5HSC7 A0A0A9Y3S2 T1JC70 A0A154PRF3 A0A131XIK6 A0A286X8N7 A0A1E1X5Z3 H3BB79 A0A0M8ZWC0 V9KCF8 L7M7J8 A0A2Y9DNE9 A0A099Z9C1 A0A0D9R101 A0A1L8HG21 A0A3P4S593 R0LXC7 A0A131YHC0 A0A091VPC2 A0A091KHZ1 A0A091W586 A0A091PMT6 A0A091RXK9 L8IUK2 E1B7N1 A0A091M183 A0A087VKA6 A0A093C5F8 A0A0A0AUC3 A0A091NTQ8 A0A091T993 A0A093NGY0 A0A094KJF8 A0A091PDJ5 G1SX62 A0A093C4S7 A0A091L6Z4 A0A091IWH3 A0A091QFX1 G3TI80 A0A1W4X0A5 I3MH61 F7HXY0 A0A093I514 U3DI47 A0A093I891 A0A384CGX9 A0A091U237 G3VE34 A0A1S3EZH0 A0A250Y397 A0A2K6BU20 A0A2K6JNC3 K9J203 G1QHR1 A0A2K5ZPZ1 F1S207 F6XXW5 A0A2R9ARL1 A0A096P4Z6 A0A2K6Q688 H0XRA2 A0A1U7SSE5
PDB
3NF1
E-value=0.000148072,
Score=109
Ontologies
GO
Topology
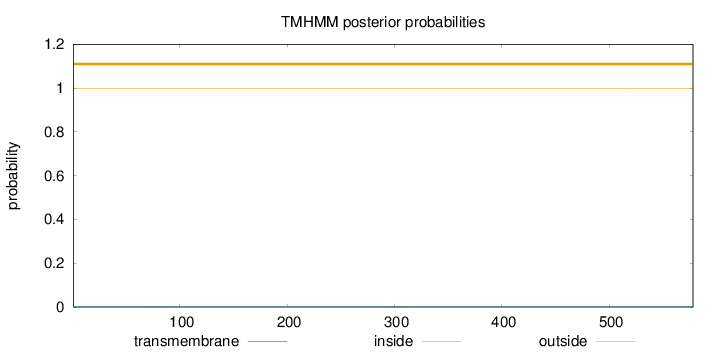
Length:
578
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0181799999999999
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00122
outside
1 - 578
Population Genetic Test Statistics
Pi
18.639224
Theta
20.303303
Tajima's D
-0.785419
CLR
1.141646
CSRT
0.179841007949603
Interpretation
Uncertain
