Pre Gene Modal
BGIBMGA009898
Annotation
PREDICTED:_protein_HID1_[Bombyx_mori]
Full name
Protein HID1
Alternative Name
Down-regulated in multiple cancers 1
HID1 domain-containing protein
Protein hid-1 homolog
HID1 domain-containing protein
Protein hid-1 homolog
Location in the cell
PlasmaMembrane Reliability : 2.457
Sequence
CDS
ATGGGTAATGCTGACACAAAATTAAACTTTCGAAAGGCGGTTGTCCAGTTAACTTCAAAGTCGCAGCCTATAGATGCGTTGGACGATAGCTTCTGGGATCAGTTCTGGTCAGAGAGCGTAACCAACGTACAAGATGTATTCGCATTGGTTCCCGGTGCGGAGATACGTGCCTTGCGCGAGGAATCTCCTAATAATTTAGCAACGCTGGTTTACAAGGCTGTGGAAAAACTCGTCAAGGTCGTGGACAGCAGCTGTAGGACGCAGAGAGAACAGCTGATCGCATTGAACTGTGCACGTCTTCTTACCAGAGTATTACCATATATGTTCGAGGAACCCGAGTGGCACAGCTTCTTTTGGTCATCGCTCCCGGCTGCAGCTGAGAATGACTCTGTACCATTAGCACAATCACTGATAAATGCCGTTTGTGATCTACTCTTCTGCCCAGACTTCACAGTAACGAGCACTAAAAGGGCAGGTCCAGATCGTGCAGAAGAGCTATCCGGCTTGGACAGCTGCGAGTATATCTGGGCTGGAGGTGTCGGCTTCGCTCGCAGTCCTCCGAGACACGCGCCCCACGAAGCTGCCAGAACCGAGCTGCTTCGTCTTCTTCTCGTCTGCTTCAGTGAAACCATATACAAGCCACCGCAGCAGGCGGCCTCACATCACAACAGATGGATCGCGTACTTCACGAGTCCGGACAACCGCCACGCCCTGCCGCTGTTCACGTCGTTGCTGAACACTGTCTGCTCTTACGACCCCGTTGGACTCGGCCTGCCGTACAACCATCTGCTTTTCGCTGACACGCTGGAGCCACTAGTTGAGGTGGCGCTTCAAATTCTCATCGTCACTCTGGACCACGACACGAGCAACGCAGTCAGCGAGGACTCCGAGGAAGTGCTGCCGGATAATCTGTTCATAAACTACTTGTCTCGCATCCACCGAGACGAGGACTTCCAGTTTGCCCTGCGAGGGGTGACCCGTCTGCTGAACAACCCGCTCGCGCAGACCTACCTGCCCAACTCCTCCAAGAAGGTCAACCTTCACCAGGAGCTGTTAGTGCTGTTCTGGAAAATGTGCGACTATAATAAGAAGTTCATGTACTACGTGCTGAAGAGCAGCGACGTGCTGGAGGTGCTGGTGCCCATACTGTACCACCTCAACGACTCCAGGGCCGACCAGTCCCGCGTCGGTCTCATGCACATCGGCGTGTTCATATTGCTTCTCTTGTCCGGGGAACGGAACCTCGGCGTGAGGCTGAACAAGCCGTACGCGGCCGCCGTGCCGATGGACATCCCGGTGTTCACGGGCACTCACGCCGACCTGCTCGTCGTGGTCTTCCACAAGATCATCACCACCGGGCACCAGCGTCTGCAGCCGCTCTTCGACTGTCTACTCACCATACTAGTTAACGTGTCGCCGTACCTCAAGACGCTGTCCATGGTCGCGTCGACCAAACTGCTGCACCTGCTGGAAGCCTTCAGCACCCCCTGGTTCCTGTTCTCGCAGCCGCACCACCACCACCTGGTCTTCTTCCTGCTGGAGATGTTCAATAACATGATACAGTATCAGTTCGATGGTAACTCCAACCTGGTGTACACGATAATACGCAAGCGCGCCGTGTTCCACAACCTGGCCAACCTGCCGCACGAGCACGCCGCCATCGCGCGCTCGCTGGCGGGCGCCGGCGGGGGCGGGGCGGCCGGGGGGGGCGCCGCGCCCGCCGCCTCCGCCCTGCCCAGGTCTCGCAGCACGGAGTCAGTGGCGGAAACTGCCATGGAAGGTTCCAGACCCGCGCAGCCTGCCGAGCCGGGGACACTTAACGCCACACTGCCCGATACACCAGCGCTGGGAACCATGACGGAGCGCGAGTCCGCACATCCGCCGGCGGCCCGGGAGCGCCTGCTGCCGGACGGGACCGAGCAGCACGACAAGCCGGACAAGACCACCGCGCCTAAGGAGAGCGGGTCGTGGCGCCCGACGGCGGAGTGGGTGGGGGGGTGGCGCGGGAAGCTCCCCCTGCAGACCATCATGAGGCTACTGCAGGTCCTCGTGCCGCAGGTCGAGAAGATATGCATCGACAAAGGTCTGACAGACGAATCGGAGATCCTCCGTTTCCTGCAGCACGGCACCCTGGTGGGGCTGCTCCCCGTGCCGCACCCCATCCTCATCAGGAAGTACCAGGCCAACGCCGGGACGGCCGCCTGGTTCCGGACTTATCTCTGGGGAGTTATCTATATACGTAACGTGGATCCTCCTATATGGTACGACACAGACGTCAAACTGTTTGAAATACAACGGGTTTAG
Protein
MGNADTKLNFRKAVVQLTSKSQPIDALDDSFWDQFWSESVTNVQDVFALVPGAEIRALREESPNNLATLVYKAVEKLVKVVDSSCRTQREQLIALNCARLLTRVLPYMFEEPEWHSFFWSSLPAAAENDSVPLAQSLINAVCDLLFCPDFTVTSTKRAGPDRAEELSGLDSCEYIWAGGVGFARSPPRHAPHEAARTELLRLLLVCFSETIYKPPQQAASHHNRWIAYFTSPDNRHALPLFTSLLNTVCSYDPVGLGLPYNHLLFADTLEPLVEVALQILIVTLDHDTSNAVSEDSEEVLPDNLFINYLSRIHRDEDFQFALRGVTRLLNNPLAQTYLPNSSKKVNLHQELLVLFWKMCDYNKKFMYYVLKSSDVLEVLVPILYHLNDSRADQSRVGLMHIGVFILLLLSGERNLGVRLNKPYAAAVPMDIPVFTGTHADLLVVVFHKIITTGHQRLQPLFDCLLTILVNVSPYLKTLSMVASTKLLHLLEAFSTPWFLFSQPHHHHLVFFLLEMFNNMIQYQFDGNSNLVYTIIRKRAVFHNLANLPHEHAAIARSLAGAGGGGAAGGGAAPAASALPRSRSTESVAETAMEGSRPAQPAEPGTLNATLPDTPALGTMTERESAHPPAARERLLPDGTEQHDKPDKTTAPKESGSWRPTAEWVGGWRGKLPLQTIMRLLQVLVPQVEKICIDKGLTDESEILRFLQHGTLVGLLPVPHPILIRKYQANAGTAAWFRTYLWGVIYIRNVDPPIWYDTDVKLFEIQRV
Summary
Description
May play an important role in the development of cancers in a broad range of tissues.
Similarity
Belongs to the hid-1 family.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Golgi apparatus
Lipoprotein
Membrane
Myristate
Phosphoprotein
Reference proteome
Feature
chain Protein HID1
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
A0A2A4JID2
A0A194PS21
A0A3S2NB89
A0A212EV59
D6WWB7
N6U4P1
+ More
R4WQV1 A0A232F4C8 K7IXD0 A0A0C9QD98 A0A2A3EIH2 A0A088APV3 A0A026WE27 A0A3L8E2K7 A0A154PE40 A0A158NEQ5 A0A1B6CIT1 E2C3V5 E2A151 E0VG76 A0A0P4VU75 A0A1Y1LDD9 A0A1W4WG61 A0A0V0G467 A0A224X7G6 A0A023FAY5 U4ULX6 T1I7V2 A0A146LHQ9 A0A1L8DB55 A0A0K8TLJ1 A0A195DZP3 A0A195FLR3 A0A0L7QLX1 F4WRP0 A0A195CRP6 A0A2H8TUD1 J9KB63 A0A1B0CR35 A0A0N0BCA7 A0A2S2QUY6 A0A1B0DCG9 C3Z701 A0A1Z5LFP3 A0A293N879 A0A2R5LCL0 A0A1S3WE04 R7VDZ0 H0XGB4 A0A3Q7VAJ2 A0A3B0J1M0 A0A1S3WE59 A0A1S2ZUJ3 A0A2Y9T5D3 A0A2Y9TB99 A0A337SF35 A0A2Y9L168 A0A2H1VPM9 A0A0F7YZ01 A0A0B8RVH7 A0A2Y9L655 H9JK47 A0A151N2U1 K9IZM1 V4B1E4 A0A340X544 M3YRN2 A0A1S3EK17 A0A340X5R7 Q08E22 A0A2K6EX28 F1RVV6 A0A1W7RBT6 A0A098M1H9 A0A1S3EIT7 F7CRE7 A0A2U3VHX4 A0A341CQE1 A0A3Q7PI80 A0A2U3Z9P1 G1KEV3 Q8IV36-2 G3QLC3 T1D774 A0A2J8Y095 U3D1C1 Q8IV36 W5N671 A0A0Q3NYU4 A0A3Q0DJJ9 U3JEU7 A0A2K6M0G6 A0A1U7UGZ6 A0A1V4KVU8 A0A0F7Z221 A0A091DEG9 A0A2K6DSV8 A0A2K5V0M4 A0A0D9QX92 A0A2K5MSR2
R4WQV1 A0A232F4C8 K7IXD0 A0A0C9QD98 A0A2A3EIH2 A0A088APV3 A0A026WE27 A0A3L8E2K7 A0A154PE40 A0A158NEQ5 A0A1B6CIT1 E2C3V5 E2A151 E0VG76 A0A0P4VU75 A0A1Y1LDD9 A0A1W4WG61 A0A0V0G467 A0A224X7G6 A0A023FAY5 U4ULX6 T1I7V2 A0A146LHQ9 A0A1L8DB55 A0A0K8TLJ1 A0A195DZP3 A0A195FLR3 A0A0L7QLX1 F4WRP0 A0A195CRP6 A0A2H8TUD1 J9KB63 A0A1B0CR35 A0A0N0BCA7 A0A2S2QUY6 A0A1B0DCG9 C3Z701 A0A1Z5LFP3 A0A293N879 A0A2R5LCL0 A0A1S3WE04 R7VDZ0 H0XGB4 A0A3Q7VAJ2 A0A3B0J1M0 A0A1S3WE59 A0A1S2ZUJ3 A0A2Y9T5D3 A0A2Y9TB99 A0A337SF35 A0A2Y9L168 A0A2H1VPM9 A0A0F7YZ01 A0A0B8RVH7 A0A2Y9L655 H9JK47 A0A151N2U1 K9IZM1 V4B1E4 A0A340X544 M3YRN2 A0A1S3EK17 A0A340X5R7 Q08E22 A0A2K6EX28 F1RVV6 A0A1W7RBT6 A0A098M1H9 A0A1S3EIT7 F7CRE7 A0A2U3VHX4 A0A341CQE1 A0A3Q7PI80 A0A2U3Z9P1 G1KEV3 Q8IV36-2 G3QLC3 T1D774 A0A2J8Y095 U3D1C1 Q8IV36 W5N671 A0A0Q3NYU4 A0A3Q0DJJ9 U3JEU7 A0A2K6M0G6 A0A1U7UGZ6 A0A1V4KVU8 A0A0F7Z221 A0A091DEG9 A0A2K6DSV8 A0A2K5V0M4 A0A0D9QX92 A0A2K5MSR2
Pubmed
26354079
22118469
18362917
19820115
23537049
23691247
+ More
28648823 20075255 24508170 30249741 21347285 20798317 20566863 27129103 28004739 25474469 26823975 26369729 21719571 18563158 28528879 23254933 17975172 25476704 19121390 22293439 19393038 30723633 26358130 19892987 11281419 14702039 15489334 17974005 21337012 21406692 22398555 23758969 25243066
28648823 20075255 24508170 30249741 21347285 20798317 20566863 27129103 28004739 25474469 26823975 26369729 21719571 18563158 28528879 23254933 17975172 25476704 19121390 22293439 19393038 30723633 26358130 19892987 11281419 14702039 15489334 17974005 21337012 21406692 22398555 23758969 25243066
EMBL
NWSH01001449
PCG71190.1
KQ459595
KPI95758.1
RSAL01000285
RVE42976.1
+ More
AGBW02012233 OWR45388.1 KQ971361 EFA08688.1 APGK01040122 KB740975 ENN76560.1 AK418056 BAN21271.1 NNAY01000980 OXU25634.1 AAZX01000018 GBYB01012369 GBYB01012370 GBYB01012372 GBYB01012373 JAG82136.1 JAG82137.1 JAG82139.1 JAG82140.1 KZ288232 PBC31510.1 KK107260 EZA54173.1 QOIP01000001 RLU26970.1 KQ434874 KZC09684.1 ADTU01013609 GEDC01023949 JAS13349.1 GL452364 EFN77341.1 GL435707 EFN72818.1 DS235132 EEB12382.1 GDKW01000378 JAI56217.1 GEZM01060792 JAV70911.1 GECL01003283 JAP02841.1 GFTR01008049 JAW08377.1 GBBI01000207 JAC18505.1 KB632267 ERL91005.1 ACPB03011620 GDHC01011031 JAQ07598.1 GFDF01010487 JAV03597.1 GDAI01002404 JAI15199.1 KQ979999 KYN18301.1 KQ981490 KYN41271.1 KQ414905 KOC59622.1 GL888292 EGI63047.1 KQ977381 KYN03167.1 GFXV01006050 MBW17855.1 ABLF02014911 AJWK01024368 AJWK01024369 KQ435922 KOX68662.1 GGMS01012364 MBY81567.1 AJVK01005059 GG666590 EEN51597.1 GFJQ02001136 JAW05834.1 GFWV01023123 MAA47850.1 GGLE01003092 MBY07218.1 AMQN01005021 KB294939 ELU13895.1 AAQR03041608 AAQR03041609 OUUW01000001 SPP75134.1 AANG04004184 ODYU01003696 SOQ42783.1 GBEW01001195 JAI09170.1 GBSH01003065 JAG65962.1 BABH01026653 AKHW03004113 KYO31124.1 GABZ01004474 JAA49051.1 KB204066 ESO82024.1 AEYP01025387 BC123459 AAI23460.1 AEMK02000080 DQIR01195318 HDB50795.1 GDAY02002877 JAV48590.1 GBSI01000381 JAC96114.1 AK074401 BC032219 BC035372 AL137556 CABD030036094 CABD030036095 CABD030036096 CABD030036097 GAAZ01002879 JAA95064.1 NDHI03003284 PNJ87715.1 GAMT01007504 GAMS01007045 GAMR01009703 GAMP01008545 JAB04357.1 JAB16091.1 JAB24229.1 JAB44210.1 AHAT01000406 AHAT01000407 LMAW01003200 KQK73451.1 AGTO01000992 LSYS01001520 OPJ88623.1 GBEX01003904 JAI10656.1 KN122402 KFO30544.1 AQIA01028585 AQIB01117583
AGBW02012233 OWR45388.1 KQ971361 EFA08688.1 APGK01040122 KB740975 ENN76560.1 AK418056 BAN21271.1 NNAY01000980 OXU25634.1 AAZX01000018 GBYB01012369 GBYB01012370 GBYB01012372 GBYB01012373 JAG82136.1 JAG82137.1 JAG82139.1 JAG82140.1 KZ288232 PBC31510.1 KK107260 EZA54173.1 QOIP01000001 RLU26970.1 KQ434874 KZC09684.1 ADTU01013609 GEDC01023949 JAS13349.1 GL452364 EFN77341.1 GL435707 EFN72818.1 DS235132 EEB12382.1 GDKW01000378 JAI56217.1 GEZM01060792 JAV70911.1 GECL01003283 JAP02841.1 GFTR01008049 JAW08377.1 GBBI01000207 JAC18505.1 KB632267 ERL91005.1 ACPB03011620 GDHC01011031 JAQ07598.1 GFDF01010487 JAV03597.1 GDAI01002404 JAI15199.1 KQ979999 KYN18301.1 KQ981490 KYN41271.1 KQ414905 KOC59622.1 GL888292 EGI63047.1 KQ977381 KYN03167.1 GFXV01006050 MBW17855.1 ABLF02014911 AJWK01024368 AJWK01024369 KQ435922 KOX68662.1 GGMS01012364 MBY81567.1 AJVK01005059 GG666590 EEN51597.1 GFJQ02001136 JAW05834.1 GFWV01023123 MAA47850.1 GGLE01003092 MBY07218.1 AMQN01005021 KB294939 ELU13895.1 AAQR03041608 AAQR03041609 OUUW01000001 SPP75134.1 AANG04004184 ODYU01003696 SOQ42783.1 GBEW01001195 JAI09170.1 GBSH01003065 JAG65962.1 BABH01026653 AKHW03004113 KYO31124.1 GABZ01004474 JAA49051.1 KB204066 ESO82024.1 AEYP01025387 BC123459 AAI23460.1 AEMK02000080 DQIR01195318 HDB50795.1 GDAY02002877 JAV48590.1 GBSI01000381 JAC96114.1 AK074401 BC032219 BC035372 AL137556 CABD030036094 CABD030036095 CABD030036096 CABD030036097 GAAZ01002879 JAA95064.1 NDHI03003284 PNJ87715.1 GAMT01007504 GAMS01007045 GAMR01009703 GAMP01008545 JAB04357.1 JAB16091.1 JAB24229.1 JAB44210.1 AHAT01000406 AHAT01000407 LMAW01003200 KQK73451.1 AGTO01000992 LSYS01001520 OPJ88623.1 GBEX01003904 JAI10656.1 KN122402 KFO30544.1 AQIA01028585 AQIB01117583
Proteomes
UP000218220
UP000053268
UP000283053
UP000007151
UP000007266
UP000019118
+ More
UP000215335 UP000002358 UP000242457 UP000005203 UP000053097 UP000279307 UP000076502 UP000005205 UP000008237 UP000000311 UP000009046 UP000192223 UP000030742 UP000015103 UP000078492 UP000078541 UP000053825 UP000007755 UP000078542 UP000007819 UP000092461 UP000053105 UP000092462 UP000001554 UP000079721 UP000014760 UP000005225 UP000286642 UP000268350 UP000248484 UP000011712 UP000248482 UP000005204 UP000050525 UP000030746 UP000265300 UP000000715 UP000081671 UP000009136 UP000233160 UP000008227 UP000002281 UP000245340 UP000252040 UP000286641 UP000001646 UP000005640 UP000001519 UP000008225 UP000018468 UP000051836 UP000189704 UP000016665 UP000233180 UP000190648 UP000028990 UP000233120 UP000233100 UP000029965 UP000233060
UP000215335 UP000002358 UP000242457 UP000005203 UP000053097 UP000279307 UP000076502 UP000005205 UP000008237 UP000000311 UP000009046 UP000192223 UP000030742 UP000015103 UP000078492 UP000078541 UP000053825 UP000007755 UP000078542 UP000007819 UP000092461 UP000053105 UP000092462 UP000001554 UP000079721 UP000014760 UP000005225 UP000286642 UP000268350 UP000248484 UP000011712 UP000248482 UP000005204 UP000050525 UP000030746 UP000265300 UP000000715 UP000081671 UP000009136 UP000233160 UP000008227 UP000002281 UP000245340 UP000252040 UP000286641 UP000001646 UP000005640 UP000001519 UP000008225 UP000018468 UP000051836 UP000189704 UP000016665 UP000233180 UP000190648 UP000028990 UP000233120 UP000233100 UP000029965 UP000233060
Pfam
PF12722 Hid1
Interpro
IPR026705
Hid-1/Ecm30
ProteinModelPortal
A0A2A4JID2
A0A194PS21
A0A3S2NB89
A0A212EV59
D6WWB7
N6U4P1
+ More
R4WQV1 A0A232F4C8 K7IXD0 A0A0C9QD98 A0A2A3EIH2 A0A088APV3 A0A026WE27 A0A3L8E2K7 A0A154PE40 A0A158NEQ5 A0A1B6CIT1 E2C3V5 E2A151 E0VG76 A0A0P4VU75 A0A1Y1LDD9 A0A1W4WG61 A0A0V0G467 A0A224X7G6 A0A023FAY5 U4ULX6 T1I7V2 A0A146LHQ9 A0A1L8DB55 A0A0K8TLJ1 A0A195DZP3 A0A195FLR3 A0A0L7QLX1 F4WRP0 A0A195CRP6 A0A2H8TUD1 J9KB63 A0A1B0CR35 A0A0N0BCA7 A0A2S2QUY6 A0A1B0DCG9 C3Z701 A0A1Z5LFP3 A0A293N879 A0A2R5LCL0 A0A1S3WE04 R7VDZ0 H0XGB4 A0A3Q7VAJ2 A0A3B0J1M0 A0A1S3WE59 A0A1S2ZUJ3 A0A2Y9T5D3 A0A2Y9TB99 A0A337SF35 A0A2Y9L168 A0A2H1VPM9 A0A0F7YZ01 A0A0B8RVH7 A0A2Y9L655 H9JK47 A0A151N2U1 K9IZM1 V4B1E4 A0A340X544 M3YRN2 A0A1S3EK17 A0A340X5R7 Q08E22 A0A2K6EX28 F1RVV6 A0A1W7RBT6 A0A098M1H9 A0A1S3EIT7 F7CRE7 A0A2U3VHX4 A0A341CQE1 A0A3Q7PI80 A0A2U3Z9P1 G1KEV3 Q8IV36-2 G3QLC3 T1D774 A0A2J8Y095 U3D1C1 Q8IV36 W5N671 A0A0Q3NYU4 A0A3Q0DJJ9 U3JEU7 A0A2K6M0G6 A0A1U7UGZ6 A0A1V4KVU8 A0A0F7Z221 A0A091DEG9 A0A2K6DSV8 A0A2K5V0M4 A0A0D9QX92 A0A2K5MSR2
R4WQV1 A0A232F4C8 K7IXD0 A0A0C9QD98 A0A2A3EIH2 A0A088APV3 A0A026WE27 A0A3L8E2K7 A0A154PE40 A0A158NEQ5 A0A1B6CIT1 E2C3V5 E2A151 E0VG76 A0A0P4VU75 A0A1Y1LDD9 A0A1W4WG61 A0A0V0G467 A0A224X7G6 A0A023FAY5 U4ULX6 T1I7V2 A0A146LHQ9 A0A1L8DB55 A0A0K8TLJ1 A0A195DZP3 A0A195FLR3 A0A0L7QLX1 F4WRP0 A0A195CRP6 A0A2H8TUD1 J9KB63 A0A1B0CR35 A0A0N0BCA7 A0A2S2QUY6 A0A1B0DCG9 C3Z701 A0A1Z5LFP3 A0A293N879 A0A2R5LCL0 A0A1S3WE04 R7VDZ0 H0XGB4 A0A3Q7VAJ2 A0A3B0J1M0 A0A1S3WE59 A0A1S2ZUJ3 A0A2Y9T5D3 A0A2Y9TB99 A0A337SF35 A0A2Y9L168 A0A2H1VPM9 A0A0F7YZ01 A0A0B8RVH7 A0A2Y9L655 H9JK47 A0A151N2U1 K9IZM1 V4B1E4 A0A340X544 M3YRN2 A0A1S3EK17 A0A340X5R7 Q08E22 A0A2K6EX28 F1RVV6 A0A1W7RBT6 A0A098M1H9 A0A1S3EIT7 F7CRE7 A0A2U3VHX4 A0A341CQE1 A0A3Q7PI80 A0A2U3Z9P1 G1KEV3 Q8IV36-2 G3QLC3 T1D774 A0A2J8Y095 U3D1C1 Q8IV36 W5N671 A0A0Q3NYU4 A0A3Q0DJJ9 U3JEU7 A0A2K6M0G6 A0A1U7UGZ6 A0A1V4KVU8 A0A0F7Z221 A0A091DEG9 A0A2K6DSV8 A0A2K5V0M4 A0A0D9QX92 A0A2K5MSR2
Ontologies
GO
PANTHER
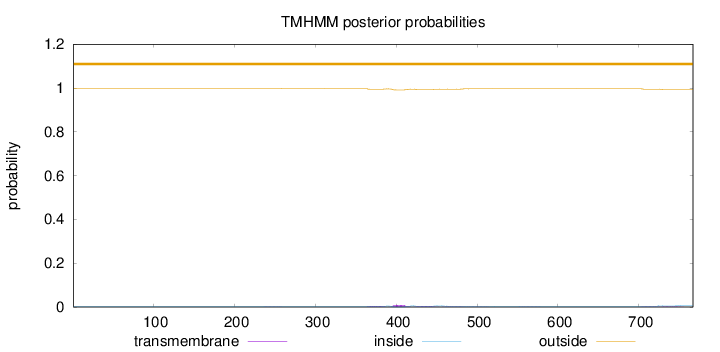
Topology
Subcellular location
Cytoplasm
Golgi apparatus membrane
Golgi apparatus membrane
Length:
767
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.47739
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00305
outside
1 - 767
Population Genetic Test Statistics
Pi
26.676817
Theta
24.311402
Tajima's D
-1.391612
CLR
18.906876
CSRT
0.0740962951852407
Interpretation
Uncertain
