Gene
KWMTBOMO04781 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009895
Annotation
PREDICTED:_6-phosphofructo-2-kinase/fructose-2?6-bisphosphatase_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.733
Sequence
CDS
ATGGATGACGAAAACCGAGCTGAAGGCTGGAAGTATTTAAATAAGCAGTTCTTCGGTGATGGAGAACGATCGAATTATGTCAACATACCGCATGTTATCGCAATGGTGGGTCTCCCGGCACGAGGGAAGACATACATTTCCAAGAAACTGTCACGCTACCTGAACTGGATAGGGATCAATACAAGAGTATTTAATCTAGGCGAATACAGAAGACATGCTACATCCGCATACACAAGCCATGAGTTCTTCAGGGCCGACAACAAAGAAGCCATGGCTATAAGACAGCAGTGCGCTTTAGATGCATTACAAGATGTGTGTGAATGGCTGACCAAAGGCGGAGAAGTCGCCGTCTTCGATGCTACAAATTCTACTTTGGACCGACGGCGGATGATAAGAGACCTTGTAGTCCACAAGATGGGCTTCAAGCTTTTCTTTGTTGAATCTATATGTGATGACCCTAGGATTATTGAACAAAACATTATGGAAGTAAAAGTGAGCAGTCCGGACTACACAAACATTCAGAACACGGATAATGTTCTAAATGATTTCATGCTCCGGATAGAACATTACAAGGAGAAGTACGAGCCTCTGGACGAGAGCCTGGAGTCTGAATACAGCTTCATGAAGATATACGATACCGGAGAGAAGGTGGTCGTTCACAAGCATGAAGGACACATACAGAGTCGGATTGTGTACTACTTGATGAACATACATATTGTACCCAGAACTATATATCTGACAAGGCACGGCGAGAGCCTGCACAACCTGGTCGGGCGCATCGGCGGTGACAGTCCGCTGTCGCCGCGCGGCTGCCAGTACGCGCGCGCGCTCTCCGCCTACATCGAGCAGCAGCGCATCCCGGGCCTGCGCGTGTGGACCTCCTGGATGCGCCGCGCCATACAGACGGTCAAGGACGTCAAGGCGCCGCAGGAGAGGTGGAAGGCCCTCAACGAGATCGACGCGGGCATATGTGAAGAGATGACATACGCTGAGATACAAGAGAAGTACCCGTCGGACTTCAACTCCCGCGACGCCAACAAGTTCGCGTACCGCTACCCCCGGGGGGAGAGCTACGAGGACCTGGTGGCCAGGCTGGAGCCCGTCATCATGGAGCTGGAGCGGCAGGGCAACGTGCTCGTGGTCTCGCACCAGGCCGTCATGCGCTGTCTGCTGGCCTACTTCCTCGACAAGAGTGCAGAGGAGCTGCCGTACCTGCAGGTGCCGCTGCACGCGGTCATCAAGCTGACGCCGGTCGCGTACGGCTGCCGCGAGGAGCACATCCGGCTCAGCGTGGACGCCGTCGACACGCACCGCCCCAAGCCGCAGATAATACCAGCGCCGCCCATAAAGCCGCCCGTGCCTCCAGTCCTGAATGGCGAAACGAATGGGGATCAAAACGACGATATATCTCATTGA
Protein
MDDENRAEGWKYLNKQFFGDGERSNYVNIPHVIAMVGLPARGKTYISKKLSRYLNWIGINTRVFNLGEYRRHATSAYTSHEFFRADNKEAMAIRQQCALDALQDVCEWLTKGGEVAVFDATNSTLDRRRMIRDLVVHKMGFKLFFVESICDDPRIIEQNIMEVKVSSPDYTNIQNTDNVLNDFMLRIEHYKEKYEPLDESLESEYSFMKIYDTGEKVVVHKHEGHIQSRIVYYLMNIHIVPRTIYLTRHGESLHNLVGRIGGDSPLSPRGCQYARALSAYIEQQRIPGLRVWTSWMRRAIQTVKDVKAPQERWKALNEIDAGICEEMTYAEIQEKYPSDFNSRDANKFAYRYPRGESYEDLVARLEPVIMELERQGNVLVVSHQAVMRCLLAYFLDKSAEELPYLQVPLHAVIKLTPVAYGCREEHIRLSVDAVDTHRPKPQIIPAPPIKPPVPPVLNGETNGDQNDDISH
Summary
Uniprot
A0A2A4JIE9
A0A2A4JHA8
A0A212EVA6
A0A0N1PFR0
A0A194PX86
A0A2A4JHE1
+ More
A0A2A4JHB8 A0A2A4JIE2 H9JK44 A0A2A4JHZ9 A0A3S2LRQ9 A0A2A4JGJ3 A0A182GE03 A0A182GNW4 A0A182S6H7 A0A182KAD2 A0A182RY36 A0A182QPR8 A0A182J9Q6 A0A182XFS1 A0A0T6BIK7 A0A182KLM8 A0A182UAG8 A0A182HHT8 W5JVA0 A0A1B6DGN2 A0A182FU81 A0A1B6CPW8 A0A1L8E1L8 A0A1B6D6F3 A0A1B0GQQ6 A0A224XPB5 A0A069DU84 A0A139WB70 Q16K24 A0A0V0G8D4 A0A182W276 Q16K23 U4UF65 A0A139WB95 Q16K25 A0A139WBA9 A0A151WLB8 U4UNE3 N6SZV0 J3JZ12 V5GRD3 V5G2C2 E2BWS4 A0A1Y1KBL5 A0A1Q3FXC4 A0A087ZWB6 A0A1Y1KBZ9 V5I8Q0 V5GVA8 A0A2A3ELF9 A0A2A3ENC1 A0A195ESF8 A0A1Y1KDY0 A0A1Y1K6G9 A0A1Q3FX18 A0A154PSC6 A0A1Q3FXF3 A0A1Q3FWZ3 A0A1Q3FWV0 A0A310SIF2 A0A151IHL5 F4X4W1 A0A232F125 A0A182N9R7 A0A2M4CUG9 A0A2J7RCS1 A0A026WG21 A0A0L7QUZ2 A0A2M4A512 A0A182LS58 A0A151HXZ5 K7J2N5 E0VK81 T1HM25 A0A2M4CIL3 A0A182VCH2 A0A0M8ZWA5 A0A1S4H937 A0A2M3Z2A3 A0A2M4A5G8 A0A2M4BM86 T1DQB8 Q7PZN8 A0A2M4BN45 A0A2M4BN49 A0A067QMV9 E2AX81 A0A1Q3FA43 A0A1Q3F934 A0A1Q3FAL9 A0A1Q3F913 A0A1Q3FAW9 A0A0K8TMX4 B0WBW5
A0A2A4JHB8 A0A2A4JIE2 H9JK44 A0A2A4JHZ9 A0A3S2LRQ9 A0A2A4JGJ3 A0A182GE03 A0A182GNW4 A0A182S6H7 A0A182KAD2 A0A182RY36 A0A182QPR8 A0A182J9Q6 A0A182XFS1 A0A0T6BIK7 A0A182KLM8 A0A182UAG8 A0A182HHT8 W5JVA0 A0A1B6DGN2 A0A182FU81 A0A1B6CPW8 A0A1L8E1L8 A0A1B6D6F3 A0A1B0GQQ6 A0A224XPB5 A0A069DU84 A0A139WB70 Q16K24 A0A0V0G8D4 A0A182W276 Q16K23 U4UF65 A0A139WB95 Q16K25 A0A139WBA9 A0A151WLB8 U4UNE3 N6SZV0 J3JZ12 V5GRD3 V5G2C2 E2BWS4 A0A1Y1KBL5 A0A1Q3FXC4 A0A087ZWB6 A0A1Y1KBZ9 V5I8Q0 V5GVA8 A0A2A3ELF9 A0A2A3ENC1 A0A195ESF8 A0A1Y1KDY0 A0A1Y1K6G9 A0A1Q3FX18 A0A154PSC6 A0A1Q3FXF3 A0A1Q3FWZ3 A0A1Q3FWV0 A0A310SIF2 A0A151IHL5 F4X4W1 A0A232F125 A0A182N9R7 A0A2M4CUG9 A0A2J7RCS1 A0A026WG21 A0A0L7QUZ2 A0A2M4A512 A0A182LS58 A0A151HXZ5 K7J2N5 E0VK81 T1HM25 A0A2M4CIL3 A0A182VCH2 A0A0M8ZWA5 A0A1S4H937 A0A2M3Z2A3 A0A2M4A5G8 A0A2M4BM86 T1DQB8 Q7PZN8 A0A2M4BN45 A0A2M4BN49 A0A067QMV9 E2AX81 A0A1Q3FA43 A0A1Q3F934 A0A1Q3FAL9 A0A1Q3F913 A0A1Q3FAW9 A0A0K8TMX4 B0WBW5
Pubmed
EMBL
NWSH01001449
PCG71203.1
PCG71199.1
AGBW02012233
OWR45391.1
KQ461192
+ More
KPJ07047.1 KQ459595 KPI95755.1 PCG71196.1 PCG71209.1 PCG71200.1 BABH01026650 PCG71204.1 RSAL01000285 RVE42981.1 PCG71205.1 JXUM01056977 JXUM01056978 KQ561934 KXJ77115.1 JXUM01077266 KQ562997 KXJ74681.1 AXCN02001804 LJIG01000213 KRT86727.1 APCN01002442 ADMH02000393 ETN66719.1 GEDC01012449 JAS24849.1 GEDC01021874 JAS15424.1 GFDF01001476 JAV12608.1 GEDC01016036 JAS21262.1 AJVK01007783 GFTR01006423 JAW10003.1 GBGD01001394 JAC87495.1 KQ971372 KYB25198.1 CH477980 EAT34650.1 GECL01001690 JAP04434.1 EAT34648.1 KB632294 ERL91677.1 KYB25197.1 EAT34649.1 KYB25196.1 KQ982974 KYQ48581.1 ERL91676.1 APGK01050297 KB741169 ENN73319.1 BT128493 AEE63450.1 GALX01004299 JAB64167.1 GALX01004301 JAB64165.1 GL451188 EFN79788.1 GEZM01091275 JAV56965.1 GFDL01002909 JAV32136.1 GEZM01091274 JAV56966.1 GALX01004300 JAB64166.1 GALX01004298 JAB64168.1 KZ288215 PBC32540.1 PBC32541.1 KQ981993 KYN31106.1 GEZM01091273 JAV56967.1 GEZM01091276 JAV56964.1 GFDL01002957 JAV32088.1 KQ435127 KZC14809.1 GFDL01002913 JAV32132.1 GFDL01002946 JAV32099.1 GFDL01002961 JAV32084.1 KQ759851 OAD62558.1 KQ977592 KYN01652.1 GL888679 EGI58534.1 NNAY01001352 OXU24269.1 GGFL01004751 MBW68929.1 NEVH01005884 PNF38625.1 KK107231 QOIP01000009 EZA55002.1 RLU19139.1 KQ414727 KOC62482.1 GGFK01002573 MBW35894.1 AXCM01001039 KQ976730 KYM76418.1 DS235239 EEB13787.1 ACPB03014353 ACPB03014354 GGFL01000803 MBW64981.1 KQ435821 KOX72420.1 AAAB01008986 GGFM01001883 MBW22634.1 GGFK01002712 MBW36033.1 GGFJ01005058 MBW54199.1 GAMD01002624 JAA98966.1 EAA00451.4 GGFJ01005057 MBW54198.1 GGFJ01005283 MBW54424.1 KK853235 KDR09557.1 GL443520 EFN61964.1 GFDL01010599 JAV24446.1 GFDL01011016 JAV24029.1 GFDL01010410 JAV24635.1 GFDL01011036 JAV24009.1 GFDL01010345 JAV24700.1 GDAI01002328 JAI15275.1 DS231882 EDS42895.1
KPJ07047.1 KQ459595 KPI95755.1 PCG71196.1 PCG71209.1 PCG71200.1 BABH01026650 PCG71204.1 RSAL01000285 RVE42981.1 PCG71205.1 JXUM01056977 JXUM01056978 KQ561934 KXJ77115.1 JXUM01077266 KQ562997 KXJ74681.1 AXCN02001804 LJIG01000213 KRT86727.1 APCN01002442 ADMH02000393 ETN66719.1 GEDC01012449 JAS24849.1 GEDC01021874 JAS15424.1 GFDF01001476 JAV12608.1 GEDC01016036 JAS21262.1 AJVK01007783 GFTR01006423 JAW10003.1 GBGD01001394 JAC87495.1 KQ971372 KYB25198.1 CH477980 EAT34650.1 GECL01001690 JAP04434.1 EAT34648.1 KB632294 ERL91677.1 KYB25197.1 EAT34649.1 KYB25196.1 KQ982974 KYQ48581.1 ERL91676.1 APGK01050297 KB741169 ENN73319.1 BT128493 AEE63450.1 GALX01004299 JAB64167.1 GALX01004301 JAB64165.1 GL451188 EFN79788.1 GEZM01091275 JAV56965.1 GFDL01002909 JAV32136.1 GEZM01091274 JAV56966.1 GALX01004300 JAB64166.1 GALX01004298 JAB64168.1 KZ288215 PBC32540.1 PBC32541.1 KQ981993 KYN31106.1 GEZM01091273 JAV56967.1 GEZM01091276 JAV56964.1 GFDL01002957 JAV32088.1 KQ435127 KZC14809.1 GFDL01002913 JAV32132.1 GFDL01002946 JAV32099.1 GFDL01002961 JAV32084.1 KQ759851 OAD62558.1 KQ977592 KYN01652.1 GL888679 EGI58534.1 NNAY01001352 OXU24269.1 GGFL01004751 MBW68929.1 NEVH01005884 PNF38625.1 KK107231 QOIP01000009 EZA55002.1 RLU19139.1 KQ414727 KOC62482.1 GGFK01002573 MBW35894.1 AXCM01001039 KQ976730 KYM76418.1 DS235239 EEB13787.1 ACPB03014353 ACPB03014354 GGFL01000803 MBW64981.1 KQ435821 KOX72420.1 AAAB01008986 GGFM01001883 MBW22634.1 GGFK01002712 MBW36033.1 GGFJ01005058 MBW54199.1 GAMD01002624 JAA98966.1 EAA00451.4 GGFJ01005057 MBW54198.1 GGFJ01005283 MBW54424.1 KK853235 KDR09557.1 GL443520 EFN61964.1 GFDL01010599 JAV24446.1 GFDL01011016 JAV24029.1 GFDL01010410 JAV24635.1 GFDL01011036 JAV24009.1 GFDL01010345 JAV24700.1 GDAI01002328 JAI15275.1 DS231882 EDS42895.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000005204
UP000283053
+ More
UP000069940 UP000249989 UP000075901 UP000075881 UP000075900 UP000075886 UP000075880 UP000076407 UP000075882 UP000075902 UP000075840 UP000000673 UP000069272 UP000092462 UP000007266 UP000008820 UP000075920 UP000030742 UP000075809 UP000019118 UP000008237 UP000005203 UP000242457 UP000078541 UP000076502 UP000078542 UP000007755 UP000215335 UP000075884 UP000235965 UP000053097 UP000279307 UP000053825 UP000075883 UP000078540 UP000002358 UP000009046 UP000015103 UP000075903 UP000053105 UP000007062 UP000027135 UP000000311 UP000002320
UP000069940 UP000249989 UP000075901 UP000075881 UP000075900 UP000075886 UP000075880 UP000076407 UP000075882 UP000075902 UP000075840 UP000000673 UP000069272 UP000092462 UP000007266 UP000008820 UP000075920 UP000030742 UP000075809 UP000019118 UP000008237 UP000005203 UP000242457 UP000078541 UP000076502 UP000078542 UP000007755 UP000215335 UP000075884 UP000235965 UP000053097 UP000279307 UP000053825 UP000075883 UP000078540 UP000002358 UP000009046 UP000015103 UP000075903 UP000053105 UP000007062 UP000027135 UP000000311 UP000002320
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JIE9
A0A2A4JHA8
A0A212EVA6
A0A0N1PFR0
A0A194PX86
A0A2A4JHE1
+ More
A0A2A4JHB8 A0A2A4JIE2 H9JK44 A0A2A4JHZ9 A0A3S2LRQ9 A0A2A4JGJ3 A0A182GE03 A0A182GNW4 A0A182S6H7 A0A182KAD2 A0A182RY36 A0A182QPR8 A0A182J9Q6 A0A182XFS1 A0A0T6BIK7 A0A182KLM8 A0A182UAG8 A0A182HHT8 W5JVA0 A0A1B6DGN2 A0A182FU81 A0A1B6CPW8 A0A1L8E1L8 A0A1B6D6F3 A0A1B0GQQ6 A0A224XPB5 A0A069DU84 A0A139WB70 Q16K24 A0A0V0G8D4 A0A182W276 Q16K23 U4UF65 A0A139WB95 Q16K25 A0A139WBA9 A0A151WLB8 U4UNE3 N6SZV0 J3JZ12 V5GRD3 V5G2C2 E2BWS4 A0A1Y1KBL5 A0A1Q3FXC4 A0A087ZWB6 A0A1Y1KBZ9 V5I8Q0 V5GVA8 A0A2A3ELF9 A0A2A3ENC1 A0A195ESF8 A0A1Y1KDY0 A0A1Y1K6G9 A0A1Q3FX18 A0A154PSC6 A0A1Q3FXF3 A0A1Q3FWZ3 A0A1Q3FWV0 A0A310SIF2 A0A151IHL5 F4X4W1 A0A232F125 A0A182N9R7 A0A2M4CUG9 A0A2J7RCS1 A0A026WG21 A0A0L7QUZ2 A0A2M4A512 A0A182LS58 A0A151HXZ5 K7J2N5 E0VK81 T1HM25 A0A2M4CIL3 A0A182VCH2 A0A0M8ZWA5 A0A1S4H937 A0A2M3Z2A3 A0A2M4A5G8 A0A2M4BM86 T1DQB8 Q7PZN8 A0A2M4BN45 A0A2M4BN49 A0A067QMV9 E2AX81 A0A1Q3FA43 A0A1Q3F934 A0A1Q3FAL9 A0A1Q3F913 A0A1Q3FAW9 A0A0K8TMX4 B0WBW5
A0A2A4JHB8 A0A2A4JIE2 H9JK44 A0A2A4JHZ9 A0A3S2LRQ9 A0A2A4JGJ3 A0A182GE03 A0A182GNW4 A0A182S6H7 A0A182KAD2 A0A182RY36 A0A182QPR8 A0A182J9Q6 A0A182XFS1 A0A0T6BIK7 A0A182KLM8 A0A182UAG8 A0A182HHT8 W5JVA0 A0A1B6DGN2 A0A182FU81 A0A1B6CPW8 A0A1L8E1L8 A0A1B6D6F3 A0A1B0GQQ6 A0A224XPB5 A0A069DU84 A0A139WB70 Q16K24 A0A0V0G8D4 A0A182W276 Q16K23 U4UF65 A0A139WB95 Q16K25 A0A139WBA9 A0A151WLB8 U4UNE3 N6SZV0 J3JZ12 V5GRD3 V5G2C2 E2BWS4 A0A1Y1KBL5 A0A1Q3FXC4 A0A087ZWB6 A0A1Y1KBZ9 V5I8Q0 V5GVA8 A0A2A3ELF9 A0A2A3ENC1 A0A195ESF8 A0A1Y1KDY0 A0A1Y1K6G9 A0A1Q3FX18 A0A154PSC6 A0A1Q3FXF3 A0A1Q3FWZ3 A0A1Q3FWV0 A0A310SIF2 A0A151IHL5 F4X4W1 A0A232F125 A0A182N9R7 A0A2M4CUG9 A0A2J7RCS1 A0A026WG21 A0A0L7QUZ2 A0A2M4A512 A0A182LS58 A0A151HXZ5 K7J2N5 E0VK81 T1HM25 A0A2M4CIL3 A0A182VCH2 A0A0M8ZWA5 A0A1S4H937 A0A2M3Z2A3 A0A2M4A5G8 A0A2M4BM86 T1DQB8 Q7PZN8 A0A2M4BN45 A0A2M4BN49 A0A067QMV9 E2AX81 A0A1Q3FA43 A0A1Q3F934 A0A1Q3FAL9 A0A1Q3F913 A0A1Q3FAW9 A0A0K8TMX4 B0WBW5
PDB
5HR5
E-value=1.8519e-153,
Score=1392
Ontologies
PATHWAY
GO
PANTHER
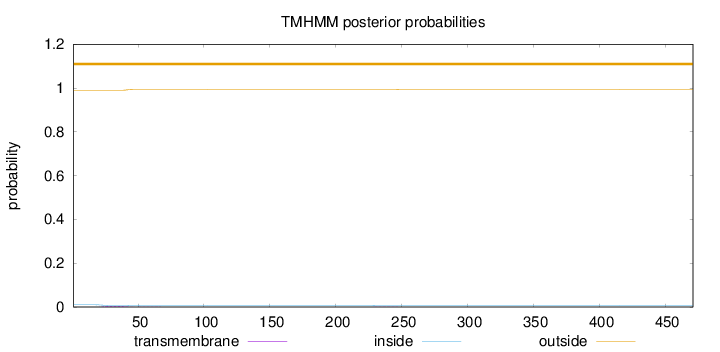
Topology
Length:
471
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08721
Exp number, first 60 AAs:
0.07278
Total prob of N-in:
0.00995
outside
1 - 471
Population Genetic Test Statistics
Pi
25.813854
Theta
25.574631
Tajima's D
0.229733
CLR
0.432216
CSRT
0.44302784860757
Interpretation
Uncertain
