Gene
KWMTBOMO04778
Pre Gene Modal
BGIBMGA009894
Annotation
PREDICTED:_Down_syndrome_cell_adhesion_molecule-like_protein_1_homolog_isoform_X1_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.442 PlasmaMembrane Reliability : 1.66
Sequence
CDS
ATGTTTTTCGGACGATTATTGTTCACTGTGTTCTGCCTTGAAATAGAAGTTTTTTCTTTCCTCGGGGCTGAAGATGTGAACATACGTGAAGTCTTTGCAGACAAGGGAACCATAAACGTAACAATACCGTGTGGGGAGCTCCTTACTGTACCACCCCCTAATGTGCAATGGGTGCACAGGGGCCACAAGACTCATCACAATGTTTTGGCTGACGGTAGTCTGCTCCTGAAGAAGATAGGAACCGAAGACGCTGGACTATACGAGTGCAGTCTTGAAAATGAGACAGATTATGTCGACAGGGTCAACTTGACCGTACGAACGGAGCCCCCGCCTCTAGTGAACGTTACCGTCCATTCCTCGACTATCCTCGCTCTGATCCTCTGGAACGTGGCCGGAGACGGTGGTCATCCCATCATAGATTTCACCGCTCAGTATAGATCGGCGATGCCAGTCAACGGGACATTGGAACCGTGGAGGCCAATCAGCCCCAATCACATTAGTCCTAATTCGCGTCAGATCGACGTGTACCATTTGGAGCCGAACGCCTCGTACTGGTTCCGAGTTTGGGCGACGAACGTGCTTGGACCGGGCCCCCCAGTCGAAGTGCTCGCTACAACCCTGTACAGTGATCAGGAAGCAGAGCTGTACAAACACTTCCTAGAGGGCGTCGAGACCTTCGACACGAGGACGTGGGTGGCGGCAGTCTGCGTGGTGATGGGCACGCTGCTGGTGCTCGCGGTCGGCACCTGCGCCGTGCTCTGCCGCGAGTGGCGACGCACCGATTACGGGGACGATCAGGATTTAATTGAACTAGTGCCTAACATCATTCTGAATCCGGGCTTCATGGAAATGGACCCGATTCGACCGCGGGAGGTGTTCGGCGGCTCGATCATCAGGTCGACGGAGTGTCCGGGCCCGGGGAAGCCGAGGAGGGTTTGA
Protein
MFFGRLLFTVFCLEIEVFSFLGAEDVNIREVFADKGTINVTIPCGELLTVPPPNVQWVHRGHKTHHNVLADGSLLLKKIGTEDAGLYECSLENETDYVDRVNLTVRTEPPPLVNVTVHSSTILALILWNVAGDGGHPIIDFTAQYRSAMPVNGTLEPWRPISPNHISPNSRQIDVYHLEPNASYWFRVWATNVLGPGPPVEVLATTLYSDQEAELYKHFLEGVETFDTRTWVAAVCVVMGTLLVLAVGTCAVLCREWRRTDYGDDQDLIELVPNIILNPGFMEMDPIRPREVFGGSIIRSTECPGPGKPRRV
Summary
Uniprot
A0A0L7KNK7
A0A0C9RVI2
A0A310SSJ9
A0A0N0BC44
E9II58
A0A2A3EPH9
+ More
V9IB04 A0A088AVS3 A0A0L7RJI4 A0A1B6MCC7 E1ZVC3 F4WJ76 A0A195CAL8 A0A151WVK8 K7IQ30 A0A151J6M4 V9I8T8 A0A158NED5 A0A195FLE3 A0A232EQ26 D6WG58 A0A1B6CI39 A0A026W7A0 A0A139WLU6 A0A154PP94 A0A146LAC8 A0A0J7KJZ0 A0A139WLH5 A0A067R238 A0A1Y1JZU3 A0A1B6L4W2 A0A023F994 A0A1B6CS62 A0A2J7PQ85 A0A1Y1JZS2 A0A0C9RMZ1 R4FK07 A0A2R7W702 A0A195BX30 A0A1W4WNG2 A0A0T6BAJ0 U4UI36 N6SXP6 A0A182M9R5 A0A226DH64 A0A1B0D4C9 A0A1S3D080 A0A1W4WCF9 A0A0Q9WD13 A0A034WUK1 A0A1B0CRF5 A0A0P8XIR6 A0A0Q5T386 M9NDT2 A0A0M3QZH2 A0A0R1EBX1 A0A087T9Z2 A0A0P5FYM6 A0A0P5EDP6 A0A2S2PUX7 A0A2H8TDB9 A0A2S2N6M2 A0A0Q9WEG6
V9IB04 A0A088AVS3 A0A0L7RJI4 A0A1B6MCC7 E1ZVC3 F4WJ76 A0A195CAL8 A0A151WVK8 K7IQ30 A0A151J6M4 V9I8T8 A0A158NED5 A0A195FLE3 A0A232EQ26 D6WG58 A0A1B6CI39 A0A026W7A0 A0A139WLU6 A0A154PP94 A0A146LAC8 A0A0J7KJZ0 A0A139WLH5 A0A067R238 A0A1Y1JZU3 A0A1B6L4W2 A0A023F994 A0A1B6CS62 A0A2J7PQ85 A0A1Y1JZS2 A0A0C9RMZ1 R4FK07 A0A2R7W702 A0A195BX30 A0A1W4WNG2 A0A0T6BAJ0 U4UI36 N6SXP6 A0A182M9R5 A0A226DH64 A0A1B0D4C9 A0A1S3D080 A0A1W4WCF9 A0A0Q9WD13 A0A034WUK1 A0A1B0CRF5 A0A0P8XIR6 A0A0Q5T386 M9NDT2 A0A0M3QZH2 A0A0R1EBX1 A0A087T9Z2 A0A0P5FYM6 A0A0P5EDP6 A0A2S2PUX7 A0A2H8TDB9 A0A2S2N6M2 A0A0Q9WEG6
Pubmed
EMBL
JTDY01008231
KOB64670.1
GBYB01012940
JAG82707.1
KQ759817
OAD62777.1
+ More
KQ435922 KOX68468.1 GL763358 EFZ19742.1 KZ288198 PBC33665.1 JR036963 AEY57504.1 KQ414582 KOC70886.1 GEBQ01006394 JAT33583.1 GL434492 EFN74833.1 GL888182 EGI65654.1 KQ978023 KYM97862.1 KQ982696 KYQ51969.1 KQ979851 KYN18855.1 JR036964 AEY57505.1 ADTU01013259 KQ981490 KYN41082.1 NNAY01002839 OXU20460.1 KQ971320 EFA00199.2 GEDC01024162 GEDC01001125 JAS13136.1 JAS36173.1 KK107388 EZA51496.1 KYB28801.1 KQ434998 KZC13274.1 GDHC01015049 JAQ03580.1 LBMM01006261 KMQ90758.1 KYB28800.1 KK853076 KDR11737.1 GEZM01099417 GEZM01099416 GEZM01099407 GEZM01099406 JAV53340.1 GEBQ01021250 JAT18727.1 GBBI01001229 JAC17483.1 GEDC01021047 GEDC01009219 GEDC01007485 JAS16251.1 JAS28079.1 JAS29813.1 NEVH01022640 PNF18499.1 GEZM01099422 GEZM01099420 GEZM01099419 JAV53320.1 GBYB01008431 JAG78198.1 ACPB03005595 ACPB03005596 ACPB03005597 GAHY01002068 JAA75442.1 KK854268 PTY13985.1 KQ976401 KYM92516.1 LJIG01002595 KRT84345.1 KB632366 ERL93644.1 APGK01052900 KB741216 ENN72529.1 AXCM01002508 LNIX01000020 OXA44037.1 AJVK01011359 AJVK01011360 AJVK01011361 AJVK01011362 AJVK01011363 AJVK01011364 AJVK01011365 AJVK01011366 CH940651 KRF82520.1 GAKP01001072 JAC57880.1 AJWK01024713 AJWK01024714 CH902635 KPU74720.1 CH954180 KQS29874.1 KQS29875.1 AE014298 AFH07317.1 CP012528 ALC49369.1 CM000162 KRK07000.1 KRK07002.1 KK114222 KFM61931.1 GDIQ01253903 JAJ97821.1 GDIP01144354 LRGB01001361 JAJ79048.1 KZS12612.1 GGMR01020515 MBY33134.1 GFXV01000279 MBW12084.1 GGMR01000151 MBY12770.1 KRF82518.1
KQ435922 KOX68468.1 GL763358 EFZ19742.1 KZ288198 PBC33665.1 JR036963 AEY57504.1 KQ414582 KOC70886.1 GEBQ01006394 JAT33583.1 GL434492 EFN74833.1 GL888182 EGI65654.1 KQ978023 KYM97862.1 KQ982696 KYQ51969.1 KQ979851 KYN18855.1 JR036964 AEY57505.1 ADTU01013259 KQ981490 KYN41082.1 NNAY01002839 OXU20460.1 KQ971320 EFA00199.2 GEDC01024162 GEDC01001125 JAS13136.1 JAS36173.1 KK107388 EZA51496.1 KYB28801.1 KQ434998 KZC13274.1 GDHC01015049 JAQ03580.1 LBMM01006261 KMQ90758.1 KYB28800.1 KK853076 KDR11737.1 GEZM01099417 GEZM01099416 GEZM01099407 GEZM01099406 JAV53340.1 GEBQ01021250 JAT18727.1 GBBI01001229 JAC17483.1 GEDC01021047 GEDC01009219 GEDC01007485 JAS16251.1 JAS28079.1 JAS29813.1 NEVH01022640 PNF18499.1 GEZM01099422 GEZM01099420 GEZM01099419 JAV53320.1 GBYB01008431 JAG78198.1 ACPB03005595 ACPB03005596 ACPB03005597 GAHY01002068 JAA75442.1 KK854268 PTY13985.1 KQ976401 KYM92516.1 LJIG01002595 KRT84345.1 KB632366 ERL93644.1 APGK01052900 KB741216 ENN72529.1 AXCM01002508 LNIX01000020 OXA44037.1 AJVK01011359 AJVK01011360 AJVK01011361 AJVK01011362 AJVK01011363 AJVK01011364 AJVK01011365 AJVK01011366 CH940651 KRF82520.1 GAKP01001072 JAC57880.1 AJWK01024713 AJWK01024714 CH902635 KPU74720.1 CH954180 KQS29874.1 KQS29875.1 AE014298 AFH07317.1 CP012528 ALC49369.1 CM000162 KRK07000.1 KRK07002.1 KK114222 KFM61931.1 GDIQ01253903 JAJ97821.1 GDIP01144354 LRGB01001361 JAJ79048.1 KZS12612.1 GGMR01020515 MBY33134.1 GFXV01000279 MBW12084.1 GGMR01000151 MBY12770.1 KRF82518.1
Proteomes
UP000037510
UP000053105
UP000242457
UP000005203
UP000053825
UP000000311
+ More
UP000007755 UP000078542 UP000075809 UP000002358 UP000078492 UP000005205 UP000078541 UP000215335 UP000007266 UP000053097 UP000076502 UP000036403 UP000027135 UP000235965 UP000015103 UP000078540 UP000192223 UP000030742 UP000019118 UP000075883 UP000198287 UP000092462 UP000079169 UP000008792 UP000092461 UP000007801 UP000008711 UP000000803 UP000092553 UP000002282 UP000054359 UP000076858
UP000007755 UP000078542 UP000075809 UP000002358 UP000078492 UP000005205 UP000078541 UP000215335 UP000007266 UP000053097 UP000076502 UP000036403 UP000027135 UP000235965 UP000015103 UP000078540 UP000192223 UP000030742 UP000019118 UP000075883 UP000198287 UP000092462 UP000079169 UP000008792 UP000092461 UP000007801 UP000008711 UP000000803 UP000092553 UP000002282 UP000054359 UP000076858
PRIDE
Interpro
IPR013783
Ig-like_fold
+ More
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR036179 Ig-like_dom_sf
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR003598 Ig_sub2
IPR013151 Immunoglobulin
IPR033027 DSCAM_chordates
IPR032983 CDO
IPR023393 START-like_dom_sf
IPR036412 HAD-like_sf
IPR031315 LNS2/PITP
IPR004177 DDHD_dom
IPR001666 PI_transfer
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR036179 Ig-like_dom_sf
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR003598 Ig_sub2
IPR013151 Immunoglobulin
IPR033027 DSCAM_chordates
IPR032983 CDO
IPR023393 START-like_dom_sf
IPR036412 HAD-like_sf
IPR031315 LNS2/PITP
IPR004177 DDHD_dom
IPR001666 PI_transfer
Gene 3D
CDD
ProteinModelPortal
A0A0L7KNK7
A0A0C9RVI2
A0A310SSJ9
A0A0N0BC44
E9II58
A0A2A3EPH9
+ More
V9IB04 A0A088AVS3 A0A0L7RJI4 A0A1B6MCC7 E1ZVC3 F4WJ76 A0A195CAL8 A0A151WVK8 K7IQ30 A0A151J6M4 V9I8T8 A0A158NED5 A0A195FLE3 A0A232EQ26 D6WG58 A0A1B6CI39 A0A026W7A0 A0A139WLU6 A0A154PP94 A0A146LAC8 A0A0J7KJZ0 A0A139WLH5 A0A067R238 A0A1Y1JZU3 A0A1B6L4W2 A0A023F994 A0A1B6CS62 A0A2J7PQ85 A0A1Y1JZS2 A0A0C9RMZ1 R4FK07 A0A2R7W702 A0A195BX30 A0A1W4WNG2 A0A0T6BAJ0 U4UI36 N6SXP6 A0A182M9R5 A0A226DH64 A0A1B0D4C9 A0A1S3D080 A0A1W4WCF9 A0A0Q9WD13 A0A034WUK1 A0A1B0CRF5 A0A0P8XIR6 A0A0Q5T386 M9NDT2 A0A0M3QZH2 A0A0R1EBX1 A0A087T9Z2 A0A0P5FYM6 A0A0P5EDP6 A0A2S2PUX7 A0A2H8TDB9 A0A2S2N6M2 A0A0Q9WEG6
V9IB04 A0A088AVS3 A0A0L7RJI4 A0A1B6MCC7 E1ZVC3 F4WJ76 A0A195CAL8 A0A151WVK8 K7IQ30 A0A151J6M4 V9I8T8 A0A158NED5 A0A195FLE3 A0A232EQ26 D6WG58 A0A1B6CI39 A0A026W7A0 A0A139WLU6 A0A154PP94 A0A146LAC8 A0A0J7KJZ0 A0A139WLH5 A0A067R238 A0A1Y1JZU3 A0A1B6L4W2 A0A023F994 A0A1B6CS62 A0A2J7PQ85 A0A1Y1JZS2 A0A0C9RMZ1 R4FK07 A0A2R7W702 A0A195BX30 A0A1W4WNG2 A0A0T6BAJ0 U4UI36 N6SXP6 A0A182M9R5 A0A226DH64 A0A1B0D4C9 A0A1S3D080 A0A1W4WCF9 A0A0Q9WD13 A0A034WUK1 A0A1B0CRF5 A0A0P8XIR6 A0A0Q5T386 M9NDT2 A0A0M3QZH2 A0A0R1EBX1 A0A087T9Z2 A0A0P5FYM6 A0A0P5EDP6 A0A2S2PUX7 A0A2H8TDB9 A0A2S2N6M2 A0A0Q9WEG6
PDB
5I99
E-value=6.9051e-07,
Score=126
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.782662
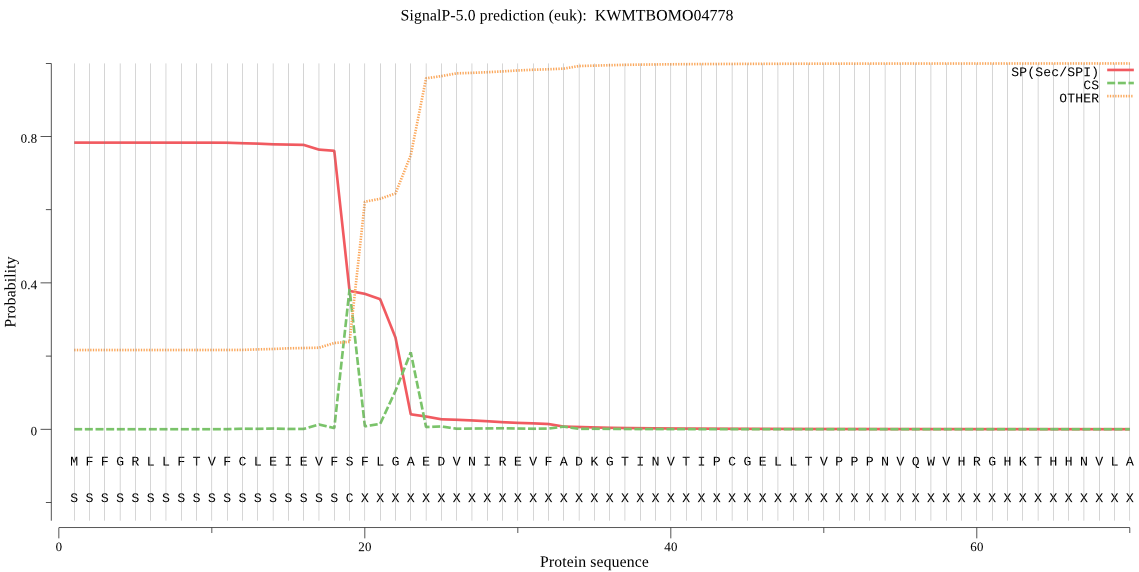
Length:
312
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.20036
Exp number, first 60 AAs:
0.12634
Total prob of N-in:
0.01894
outside
1 - 230
TMhelix
231 - 253
inside
254 - 312

Population Genetic Test Statistics
Pi
24.426129
Theta
23.018966
Tajima's D
0
CLR
1.703882
CSRT
0.370081495925204
Interpretation
Uncertain
