Gene
KWMTBOMO04770
Pre Gene Modal
BGIBMGA009850
Annotation
PREDICTED:_uncharacterized_protein_LOC101743016_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.826
Sequence
CDS
ATGGCAAAGGAAATGATGATAGTATTTGTGTACGACACACAATGTTGTACAGCTGAAGAGGATGATCCCATAAATGCTGTGCTGTATTTCCACCCAGGATGGGTGTCTGACACTCAAAGACATGCTCTAGCAGGACAGGTGGTTGGAGCAGCTCATTGCGTCAAAACGTTGCTATCTCCGCCGGTTTCAATCACCCTGCAGAGCGGAAAGTTCATCATCAGAGAATACGGAAGATACGTATTGGCGATAGGCAGCGACCGAAACATCCCAGATTGGGTGTTGAAGAATCGCGCCAACCTCCTGACCTCAATGATCAAAGTGTACCACGGCGACCTGCAGTGCTTGGCCCAGCAAATGGAAGACCAGAAACGGCTCGCCGAGAAACTGTACCAAGTTTTTGAGACTTACCTACCGGTTCTTCAGTACGGGTGCCACATATTCCAGAGGGTTCCAATGCTTAGTCTCCCGAAGAGCGCCACGTCCACTTACATGGAGTCGATGCAGATACTGGAACATTGTCGCAAAAGTACCGGTGTACTCGGCGGCGTAATACTTTACAATAACAAGATCATAGCCACACAGCTGCCCCCGGGTCTGACGTCATACCTCACCGTGGTCGACCCCTACCGGATAAAGTCCCCAGCTGAGACTCTCGAAACCGAAGCTCCCCTGCCGCTGGGGGCTCAGCTCCTCGTTGTGTACGTCAGGAGGAAGCTGTACGACAACCTCAAGAGACAGACTGAGAAATTGCAAGAGTTTTATCAAAACGGAGAAGATATTATTGCCAGGTTCAAAAAGAATCAAGAGGCGGAACGAGAAAGGCAACGTGAGTTCCCCCAGACGGGAATGAAGAGAGACAAGTCGTTACTGTTCACGGCCGTCCCGGAGGAAGACCACACAATGATCTCTCCGCCGAATCGAGAAGAGATCCCCACCGCTGAGCACCGCAAGGCCAGCGTGCCCGACGTGGTCCCGTTCACGAACAAGCCCCGTCCCAGGCCCAACAAGCTCTCGCTGAGCTTCAAGACCCAGCAGTCACTGGACGAAGACGCCAAAGAGAACGATAAAGTTTTCACGGGACAAACCAGCGTGTGCTCCACGCCGATGCTGGAGTACAAGCGCCTCCACGGGAACATGCTCTCCATCTGCCAGAACCCGGACACGAACTACCAAGAGAAGCCGATCGAGCCGGACGTGTTGAAGAACATCGAGAACTGCACGAAGATCAACTTCGACAACGAGAAGCGCGACAACCAAATCGTTCTGGACATGAACTGCATCGCGGACCACTACATCAACAAGCCGCAGCCCATCCGCAAGCCGGCCAGCGTCACGGACATACAGAGCCTCCCCCAGAGAGCCGCGTCCAAGTTCAAGCTGACAAAGTCCCACAACCCCCCGGACGACGACACCCCGTCTCCGGACGTCGACTCGCTGAAGAGCCAGTGCACCATGACGATAACCGATCCGTCCTTCCCGGTGTTCAGGAACGACGGCGTCGCCATATCGGAGTCGCTCTTCAACCAGTACGTGGAACAGTATTATTCGAGAATGAAGCAGGACTCGAAGGAGCCGAACATGTTTTCGTTCAACACGAAGCTGTGCGAGCAAGACAGGTTCGACGACTTCGATTCGGAACTGCACAGGTCTCCGCAGAGGACGCCCAAGAAATTCGCTAAGGATCCCGCCAAATCACTCTCCACAGACCATTCCAGAAGGAAATCTCTCTCGCTGCCGCTGAAGTCTCTATCCGAAAGTTCGGACTCTCAAATCGGCTCTGACTCTGAAGCGACTAGTTTCAAGAAAAAACTCACCGGCGTCCAACTCACTCCCCTGATGGAGAAACTGAGTCATCTAGCATTCTCTGACAAGTCAAGCGGATACAGCAGCAGAGTCATGACGCCTTTAGAACTGAGAGAATTCTCCACTCCGGCCGCAGAGAGGCAGATCACGTTTTGTGATAGGCCAAAAACTCAACGCGTCGAGTGTGATAGCGACGAAGACGACGACCTGGACATTTTACCGGAGTACAGCTCGCATGCCGTTAAGTGCGCCCTGTTCGTTAGTGGACTGCAGAACATGGCGCTGCTGGCGTTACTGGACATTGAGGCCGCGAATGATGCCGATACTATCAATTCACTGTGGGAAACATCCCTGAACGCGTTGGGCCCTATCGAACAGAAGTGCATGGAACCACTCGGAGCGGGCGGCGAGACTTCAGACTACAGCTACCTGCTGTTAGATCCAGACTGGGGAACTGTCAATAAGGGAGGTCCCTGGGCAGCAACAGACATCGCCACTATGGGCCTCATACACAATGACTTCTCCGACGATCCTGAACTCTCCGAACTAATACTAAGGAGCGACGAGAGCGTGGTGGTGGGCGCCAGCAGCGGCGGCGCGGAGGTGTACTACTGCGAGCGGGGCCCGCGCGACGCCGGCCCGCCCCCGCCCTCCGACGCGCTGGCGGCCGCCCCGCTGCGGGCGCGGCGCCGCCTGCACCGGGACCACGCCACGCTGCTGCTCTAA
Protein
MAKEMMIVFVYDTQCCTAEEDDPINAVLYFHPGWVSDTQRHALAGQVVGAAHCVKTLLSPPVSITLQSGKFIIREYGRYVLAIGSDRNIPDWVLKNRANLLTSMIKVYHGDLQCLAQQMEDQKRLAEKLYQVFETYLPVLQYGCHIFQRVPMLSLPKSATSTYMESMQILEHCRKSTGVLGGVILYNNKIIATQLPPGLTSYLTVVDPYRIKSPAETLETEAPLPLGAQLLVVYVRRKLYDNLKRQTEKLQEFYQNGEDIIARFKKNQEAERERQREFPQTGMKRDKSLLFTAVPEEDHTMISPPNREEIPTAEHRKASVPDVVPFTNKPRPRPNKLSLSFKTQQSLDEDAKENDKVFTGQTSVCSTPMLEYKRLHGNMLSICQNPDTNYQEKPIEPDVLKNIENCTKINFDNEKRDNQIVLDMNCIADHYINKPQPIRKPASVTDIQSLPQRAASKFKLTKSHNPPDDDTPSPDVDSLKSQCTMTITDPSFPVFRNDGVAISESLFNQYVEQYYSRMKQDSKEPNMFSFNTKLCEQDRFDDFDSELHRSPQRTPKKFAKDPAKSLSTDHSRRKSLSLPLKSLSESSDSQIGSDSEATSFKKKLTGVQLTPLMEKLSHLAFSDKSSGYSSRVMTPLELREFSTPAAERQITFCDRPKTQRVECDSDEDDDLDILPEYSSHAVKCALFVSGLQNMALLALLDIEAANDADTINSLWETSLNALGPIEQKCMEPLGAGGETSDYSYLLLDPDWGTVNKGGPWAATDIATMGLIHNDFSDDPELSELILRSDESVVVGASSGGAEVYYCERGPRDAGPPPPSDALAAAPLRARRRLHRDHATLLL
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. rRNA adenine N(6)-methyltransferase family.
Uniprot
H9JJZ9
A0A2H1WTM8
A0A2A4ISF5
A0A3S2LI72
A0A194PX75
A0A0N1I4P2
+ More
A0A212ER15 A0A2A4ITY8 A0A0L7LL31 A0A1Y1K6Y7 A0A0M8ZUG7 D7EJH5 A0A154PCZ0 A0A139W8C3 A0A0L7QZ17 F4WEM3 A0A158NF21 A0A0C9RFL7 A0A0C9RN59 A0A1I8MGY0 T1PJ06 A0A1I8MGY5 A0A1I8MGX7 A0A151WHU9 A0A1A9YCY9 A0A195E7Z8 A0A1A9VQB1 A0A195AUY4 A0A1B0FHV8 A0A1A9ZAH1 A0A195FL09 Q16R21 A0A1B0C1U0 A0A0Q5VKQ7 A0A1W4V0Q6 A0A195C456 Q8MT61 A1ZAX6 B3NML6 A0A0R1DS70 A0A1Q3G3P5 A0A1A9WC71 A0A1W4VD16 A0A0J9RFA2 A1ZAX8 A0A0P8XPK7 B4P556 B3MIE5 A0A0A1WJ60 B4QBC2 A0A0J9RFH7 A0A0K8UTM1 B4MRZ3 B4J6C4 A0A1B0D979 B4HMY9 A0A0K8WB22 A0A1I8Q8B8 A0A0K8UP90 A0A1I8Q8F1 A0A1I8Q8B4 A0A026WR63 A0A1B0GIU5 A0A0Q9WEH7 A0A1I8Q8B6 A0A0K8UPT4 B4LM62 A0A1J1I337 A0A0Q9XL59 A0A3B0JJ62 B4KNP5 A0A0M4EUA7 A0A0R3NNN9 Q28Z46 B4GIA6 A0A336M0T0 A0A1Y1JZG2 A0A182Q042 A0A182W3D5
A0A212ER15 A0A2A4ITY8 A0A0L7LL31 A0A1Y1K6Y7 A0A0M8ZUG7 D7EJH5 A0A154PCZ0 A0A139W8C3 A0A0L7QZ17 F4WEM3 A0A158NF21 A0A0C9RFL7 A0A0C9RN59 A0A1I8MGY0 T1PJ06 A0A1I8MGY5 A0A1I8MGX7 A0A151WHU9 A0A1A9YCY9 A0A195E7Z8 A0A1A9VQB1 A0A195AUY4 A0A1B0FHV8 A0A1A9ZAH1 A0A195FL09 Q16R21 A0A1B0C1U0 A0A0Q5VKQ7 A0A1W4V0Q6 A0A195C456 Q8MT61 A1ZAX6 B3NML6 A0A0R1DS70 A0A1Q3G3P5 A0A1A9WC71 A0A1W4VD16 A0A0J9RFA2 A1ZAX8 A0A0P8XPK7 B4P556 B3MIE5 A0A0A1WJ60 B4QBC2 A0A0J9RFH7 A0A0K8UTM1 B4MRZ3 B4J6C4 A0A1B0D979 B4HMY9 A0A0K8WB22 A0A1I8Q8B8 A0A0K8UP90 A0A1I8Q8F1 A0A1I8Q8B4 A0A026WR63 A0A1B0GIU5 A0A0Q9WEH7 A0A1I8Q8B6 A0A0K8UPT4 B4LM62 A0A1J1I337 A0A0Q9XL59 A0A3B0JJ62 B4KNP5 A0A0M4EUA7 A0A0R3NNN9 Q28Z46 B4GIA6 A0A336M0T0 A0A1Y1JZG2 A0A182Q042 A0A182W3D5
Pubmed
EMBL
BABH01026625
BABH01026626
ODYU01010969
SOQ56411.1
NWSH01008327
PCG62599.1
+ More
RSAL01000005 RVE54380.1 KQ459595 KPI95745.1 KQ461192 KPJ07037.1 AGBW02013148 OWR43938.1 PCG62600.1 JTDY01000710 KOB76135.1 GEZM01096818 JAV54557.1 KQ435840 KOX71400.1 KQ973453 KXZ75527.1 KQ434874 KZC09693.1 KXZ75526.1 KQ414685 KOC63807.1 GL888103 EGI67386.1 ADTU01013709 GBYB01005761 JAG75528.1 GBYB01003711 GBYB01008516 JAG73478.1 JAG78283.1 KA647898 AFP62527.1 KQ983106 KYQ47413.1 KQ979479 KYN21328.1 KQ976736 KYM76053.1 CCAG010015795 KQ981490 KYN41355.1 CH477726 EAT36853.1 JXJN01024172 CH954179 KQS62038.1 KQ978287 KYM95642.1 AY118362 AAM48391.1 AE013599 BT050428 AAG22260.1 ACJ13135.1 EDV54955.1 CM000158 KRK00064.1 GFDL01000659 JAV34386.1 CM002911 KMY94652.1 AAF57814.1 CH902619 KPU76554.1 EDW91757.1 EDV36993.1 GBXI01015591 JAC98700.1 CM000362 EDX07546.1 KMY94650.1 KMY94651.1 GDHF01022270 JAI30044.1 CH963850 EDW74882.2 CH916367 EDW00897.1 AJVK01004272 CH480816 EDW48339.1 GDHF01003921 JAI48393.1 GDHF01023812 JAI28502.1 KK107119 EZA58525.1 AJWK01017657 CH940648 KRF79679.1 GDHF01023791 JAI28523.1 EDW60940.1 CVRI01000035 CRK92785.1 CH933808 KRG05031.1 OUUW01000001 SPP73266.1 EDW10030.1 CP012524 ALC40970.1 CM000071 KRT02532.1 EAL25769.2 CH479183 EDW36226.1 UFQT01000257 SSX22443.1 GEZM01096820 JAV54553.1 AXCN02000234
RSAL01000005 RVE54380.1 KQ459595 KPI95745.1 KQ461192 KPJ07037.1 AGBW02013148 OWR43938.1 PCG62600.1 JTDY01000710 KOB76135.1 GEZM01096818 JAV54557.1 KQ435840 KOX71400.1 KQ973453 KXZ75527.1 KQ434874 KZC09693.1 KXZ75526.1 KQ414685 KOC63807.1 GL888103 EGI67386.1 ADTU01013709 GBYB01005761 JAG75528.1 GBYB01003711 GBYB01008516 JAG73478.1 JAG78283.1 KA647898 AFP62527.1 KQ983106 KYQ47413.1 KQ979479 KYN21328.1 KQ976736 KYM76053.1 CCAG010015795 KQ981490 KYN41355.1 CH477726 EAT36853.1 JXJN01024172 CH954179 KQS62038.1 KQ978287 KYM95642.1 AY118362 AAM48391.1 AE013599 BT050428 AAG22260.1 ACJ13135.1 EDV54955.1 CM000158 KRK00064.1 GFDL01000659 JAV34386.1 CM002911 KMY94652.1 AAF57814.1 CH902619 KPU76554.1 EDW91757.1 EDV36993.1 GBXI01015591 JAC98700.1 CM000362 EDX07546.1 KMY94650.1 KMY94651.1 GDHF01022270 JAI30044.1 CH963850 EDW74882.2 CH916367 EDW00897.1 AJVK01004272 CH480816 EDW48339.1 GDHF01003921 JAI48393.1 GDHF01023812 JAI28502.1 KK107119 EZA58525.1 AJWK01017657 CH940648 KRF79679.1 GDHF01023791 JAI28523.1 EDW60940.1 CVRI01000035 CRK92785.1 CH933808 KRG05031.1 OUUW01000001 SPP73266.1 EDW10030.1 CP012524 ALC40970.1 CM000071 KRT02532.1 EAL25769.2 CH479183 EDW36226.1 UFQT01000257 SSX22443.1 GEZM01096820 JAV54553.1 AXCN02000234
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000053105 UP000007266 UP000076502 UP000053825 UP000007755 UP000005205 UP000095301 UP000075809 UP000092443 UP000078492 UP000078200 UP000078540 UP000092444 UP000092445 UP000078541 UP000008820 UP000092460 UP000008711 UP000192221 UP000078542 UP000000803 UP000002282 UP000091820 UP000007801 UP000000304 UP000007798 UP000001070 UP000092462 UP000001292 UP000095300 UP000053097 UP000092461 UP000008792 UP000183832 UP000009192 UP000268350 UP000092553 UP000001819 UP000008744 UP000075886 UP000075920
UP000037510 UP000053105 UP000007266 UP000076502 UP000053825 UP000007755 UP000005205 UP000095301 UP000075809 UP000092443 UP000078492 UP000078200 UP000078540 UP000092444 UP000092445 UP000078541 UP000008820 UP000092460 UP000008711 UP000192221 UP000078542 UP000000803 UP000002282 UP000091820 UP000007801 UP000000304 UP000007798 UP000001070 UP000092462 UP000001292 UP000095300 UP000053097 UP000092461 UP000008792 UP000183832 UP000009192 UP000268350 UP000092553 UP000001819 UP000008744 UP000075886 UP000075920
Interpro
IPR026091
HPS4
+ More
IPR018501 DDT_dom
IPR019787 Znf_PHD-finger
IPR018359 Bromodomain_CS
IPR036427 Bromodomain-like_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR001487 Bromodomain
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR028942 WHIM1_dom
IPR038028 BPTF
IPR011011 Znf_FYVE_PHD
IPR028941 WHIM2_dom
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR029063 SAM-dependent_MTases
IPR001737 KsgA/Erm
IPR020596 rRNA_Ade_Mease_Trfase_CS
IPR011530 rRNA_adenine_dimethylase
IPR020598 rRNA_Ade_methylase_Trfase_N
IPR018501 DDT_dom
IPR019787 Znf_PHD-finger
IPR018359 Bromodomain_CS
IPR036427 Bromodomain-like_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR001487 Bromodomain
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR028942 WHIM1_dom
IPR038028 BPTF
IPR011011 Znf_FYVE_PHD
IPR028941 WHIM2_dom
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR029063 SAM-dependent_MTases
IPR001737 KsgA/Erm
IPR020596 rRNA_Ade_Mease_Trfase_CS
IPR011530 rRNA_adenine_dimethylase
IPR020598 rRNA_Ade_methylase_Trfase_N
Gene 3D
ProteinModelPortal
H9JJZ9
A0A2H1WTM8
A0A2A4ISF5
A0A3S2LI72
A0A194PX75
A0A0N1I4P2
+ More
A0A212ER15 A0A2A4ITY8 A0A0L7LL31 A0A1Y1K6Y7 A0A0M8ZUG7 D7EJH5 A0A154PCZ0 A0A139W8C3 A0A0L7QZ17 F4WEM3 A0A158NF21 A0A0C9RFL7 A0A0C9RN59 A0A1I8MGY0 T1PJ06 A0A1I8MGY5 A0A1I8MGX7 A0A151WHU9 A0A1A9YCY9 A0A195E7Z8 A0A1A9VQB1 A0A195AUY4 A0A1B0FHV8 A0A1A9ZAH1 A0A195FL09 Q16R21 A0A1B0C1U0 A0A0Q5VKQ7 A0A1W4V0Q6 A0A195C456 Q8MT61 A1ZAX6 B3NML6 A0A0R1DS70 A0A1Q3G3P5 A0A1A9WC71 A0A1W4VD16 A0A0J9RFA2 A1ZAX8 A0A0P8XPK7 B4P556 B3MIE5 A0A0A1WJ60 B4QBC2 A0A0J9RFH7 A0A0K8UTM1 B4MRZ3 B4J6C4 A0A1B0D979 B4HMY9 A0A0K8WB22 A0A1I8Q8B8 A0A0K8UP90 A0A1I8Q8F1 A0A1I8Q8B4 A0A026WR63 A0A1B0GIU5 A0A0Q9WEH7 A0A1I8Q8B6 A0A0K8UPT4 B4LM62 A0A1J1I337 A0A0Q9XL59 A0A3B0JJ62 B4KNP5 A0A0M4EUA7 A0A0R3NNN9 Q28Z46 B4GIA6 A0A336M0T0 A0A1Y1JZG2 A0A182Q042 A0A182W3D5
A0A212ER15 A0A2A4ITY8 A0A0L7LL31 A0A1Y1K6Y7 A0A0M8ZUG7 D7EJH5 A0A154PCZ0 A0A139W8C3 A0A0L7QZ17 F4WEM3 A0A158NF21 A0A0C9RFL7 A0A0C9RN59 A0A1I8MGY0 T1PJ06 A0A1I8MGY5 A0A1I8MGX7 A0A151WHU9 A0A1A9YCY9 A0A195E7Z8 A0A1A9VQB1 A0A195AUY4 A0A1B0FHV8 A0A1A9ZAH1 A0A195FL09 Q16R21 A0A1B0C1U0 A0A0Q5VKQ7 A0A1W4V0Q6 A0A195C456 Q8MT61 A1ZAX6 B3NML6 A0A0R1DS70 A0A1Q3G3P5 A0A1A9WC71 A0A1W4VD16 A0A0J9RFA2 A1ZAX8 A0A0P8XPK7 B4P556 B3MIE5 A0A0A1WJ60 B4QBC2 A0A0J9RFH7 A0A0K8UTM1 B4MRZ3 B4J6C4 A0A1B0D979 B4HMY9 A0A0K8WB22 A0A1I8Q8B8 A0A0K8UP90 A0A1I8Q8F1 A0A1I8Q8B4 A0A026WR63 A0A1B0GIU5 A0A0Q9WEH7 A0A1I8Q8B6 A0A0K8UPT4 B4LM62 A0A1J1I337 A0A0Q9XL59 A0A3B0JJ62 B4KNP5 A0A0M4EUA7 A0A0R3NNN9 Q28Z46 B4GIA6 A0A336M0T0 A0A1Y1JZG2 A0A182Q042 A0A182W3D5
Ontologies
GO
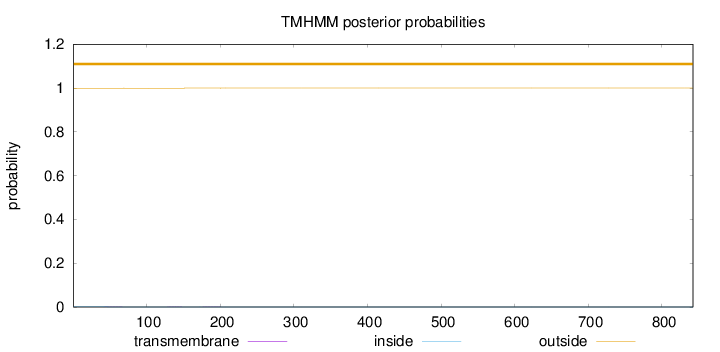
Topology
Length:
842
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02038
Exp number, first 60 AAs:
0.00627
Total prob of N-in:
0.00094
outside
1 - 842
Population Genetic Test Statistics
Pi
27.304835
Theta
25.162762
Tajima's D
0.46902
CLR
0.668315
CSRT
0.506024698765062
Interpretation
Uncertain
