Gene
KWMTBOMO04766
Pre Gene Modal
BGIBMGA009887
Annotation
PREDICTED:_leucine-rich_repeat_G_protein-coupled_receptor_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.491
Sequence
CDS
ATGAGTTCATTGAACTACAACCTGATCGAGGAAGTTCCGAAGCATGCCTTCAAAAACGCGCAAATTTCAAAACTGAGCTTCAAAGGGAACACTAAGCTGAAACGCTTGGATGAGCATGCGTTTGCGGGAAATTTACTATTACGACAGCTAGATTTAAGTAACACAGCCATTACCTCGCTACCCACTAAAGGACTTGAGAAACTGCAAATACTCAGAATTGAAAGGACTCCATCATTGAAATACATACCATCGATTTACGAATTTCAGCAACTGGAGAAAGCGTACCTGACCCATCACTTCCACTGCTGCGCGTTCAAGTTCCCGGAGATACACAATCCCGCCAGACACAAATTATACGAGACACAGATGGCGATGATGATGCAGAGATGTGCCAGTATACAAAAGTCGCAGGCTCGGAAGCGAAGGTCCTTGGAGCCGATCCGACCCGTCACGGACGGAGCGCAGACTGTCACTGCATTAGAAGATGATGATGCATCTACAATGACTGCGAGCGAAGAATATGAAAACTTCGAAGAGTACTTCTCGGACTCTGGCAGTTTCGAAGATGACGATCAAGGAGAATTCCACGACATTGTCAATGACACTGTCGTGTCGATATCTGCTGACTGTGGCAATTTCAACACAAGAAACCGGAACGTGGAATGCACTCCAGCATCCGACGCCCTGAACCCGTGCGAGGACGTGATGGGCTGGTCGTGGCTGCGGGCGAGCGTGTGGGTGGTGGTGGCGGCCGCGGTGGTGGGGAACGTGGCCGTGCTCCTCGTGCTCCTCACCAACCACACGGAGCTCACCGTCCCGCGGTTCCTCATGTGCAACCTCGCCTTCTCCGATCTCTGCACGGGACTGTACCTCCTCATGCTGGCCGTCGTCGACTTGAGGTCCTACGGGGAGTTCTTCAACTACGCCTACAACTGGCAGTACGGCGTCGGCTGCAAGATAGCCGGTTTCTTGTCAGTTTTCTCCGGGCAACTCTCGGTCATCACCTTGACGATTGTCACGCTGGAAAGGTGGTTCGCGATCACGTACGCCATTTATTTGGAAAGAAGGATTTCTTTGTCGACAGCAGCGAAAATTATGCTCGGAGGGTGGCTGTTCTCTTCGTTGATGGCGGGGCTGCCGTTACTCGGAGTCTCTGACTATTCATCGACTTCGATTTGTTTGCCGGTCGAGAGTAAAGACATAGGGGACGTGATTTATCAAGGGTCTTTGTTCCTGACCAACGCGTTGGCGTGGGTCACGATTGTCGTGTGCTACGTCCAGATCTACCGATCGTTGGGCGGTGGGGGGGAGAATTATGGGGGGAGGCGAGCCGCCGCGGCTGCTGAGCGGCGGATAGCCAACAAAATGGCTCTGCTCATAGGCACAGATTTGCTTTGCTGGGCGCCCGTCGCCTTCTTCGGGGTCACAGCCCTGGCGGGAGTGCCACTGGTCGACGTCAGTCACGGGAAAGTTTTACTAGTGTTCTTTTACCCGTTGAACGCTTGCGCCAATCCGTTCTTGTACGCGATCCTGACAAAGCAGTACCGGCGGGACTTCATAACGCTGGTAGCGAGGACGGGTCAATGTAACTGGTTGGTCGAGAAGTACAAGCTATCGACGACCCCCCCTCCGACCGCGCACACCAACCCCTCGACGCCGGCACAGCTGATGCCCCTGGTAGACCAGAAGAATCATTCGCAAGTCAGTAAAGAGTTTAGCGATTTTAAAGCGTAA
Protein
MSSLNYNLIEEVPKHAFKNAQISKLSFKGNTKLKRLDEHAFAGNLLLRQLDLSNTAITSLPTKGLEKLQILRIERTPSLKYIPSIYEFQQLEKAYLTHHFHCCAFKFPEIHNPARHKLYETQMAMMMQRCASIQKSQARKRRSLEPIRPVTDGAQTVTALEDDDASTMTASEEYENFEEYFSDSGSFEDDDQGEFHDIVNDTVVSISADCGNFNTRNRNVECTPASDALNPCEDVMGWSWLRASVWVVVAAAVVGNVAVLLVLLTNHTELTVPRFLMCNLAFSDLCTGLYLLMLAVVDLRSYGEFFNYAYNWQYGVGCKIAGFLSVFSGQLSVITLTIVTLERWFAITYAIYLERRISLSTAAKIMLGGWLFSSLMAGLPLLGVSDYSSTSICLPVESKDIGDVIYQGSLFLTNALAWVTIVVCYVQIYRSLGGGGENYGGRRAAAAAERRIANKMALLIGTDLLCWAPVAFFGVTALAGVPLVDVSHGKVLLVFFYPLNACANPFLYAILTKQYRRDFITLVARTGQCNWLVEKYKLSTTPPPTAHTNPSTPAQLMPLVDQKNHSQVSKEFSDFKA
Summary
Uniprot
Q86S10
B6VCU5
A0A2H1WJT0
A0A0N1PER8
A0A212F6E5
A0A0L7LJZ7
+ More
A0A2A4JJ49 A0A0S1YDB2 A0A194PX70 H9JK36 A0A1J1HGD8 A0A2P8XE68 A0A067QID7 A0A182YF55 A0A182M4B0 A0A084VTJ9 A0A182L7T4 A0A182I861 Q7PSL6 A0A182XFE1 A0A182VX05 A0A182RSK6 Q17CY4 A0A182P438 W8PGR8 A0A182TIN3 A0A2J7QDG0 A0A139WFY6 D6WU57 A0A067QJD2 A0A182NG31 A0A182J075 A0A182FA27 N1NVE8 A0A2U9PFW7 A0A336M4P4 A0A1W4V5Q2 A0A1B6LGF2 E0VUL7 X1WK10 B4QV48 Q9VEG4 B3MTN6 Q94979 B3P0G8 A0A1B6M0P2 B4IBD8 A0A2S2PQ43 A0A2H8TYZ0 B0WKK1 B4PKI1 A0A2S2QQ37 Q8SX01 B4NFK8 U3U984 A0A336M7W2 B4JUG0 A0A1B6E925 A0A3B0K2J6 B5DV37 B5DW24 B4KDQ2 B4GMH9 A0A1A9WBU3 A0A1B0B1Q2 A0A0Q9WBH2 B4M5G8 A0A1W4W6I0 A0A2U9PG29 T1HT73 A0A1A9XYP8 A0A0R3NK35 A0A1B6FIP4 A0A1B6H2E8 A0A1B6LJ02 A0A1B6MF70 A0A1B6KV52 A0A2R7WBA7 D6WSS0 A0A0M4EIV1 A0A1B0A9S3 A0A182K8C9 A0A0K8UNB6 W5JNP0 A0A0K8VSY6 U3UA09 A0A0P6IQ05 E9HAA1 A0A0K8UBQ4 A0A034WB40 V3ZKN4 W8C5K0 A0A139WFA3 J9JW42 K1QUZ3 A0A2T7NZD4 A0A0P5NUL5 A0A2P8YYD8 A0A210PHD1
A0A2A4JJ49 A0A0S1YDB2 A0A194PX70 H9JK36 A0A1J1HGD8 A0A2P8XE68 A0A067QID7 A0A182YF55 A0A182M4B0 A0A084VTJ9 A0A182L7T4 A0A182I861 Q7PSL6 A0A182XFE1 A0A182VX05 A0A182RSK6 Q17CY4 A0A182P438 W8PGR8 A0A182TIN3 A0A2J7QDG0 A0A139WFY6 D6WU57 A0A067QJD2 A0A182NG31 A0A182J075 A0A182FA27 N1NVE8 A0A2U9PFW7 A0A336M4P4 A0A1W4V5Q2 A0A1B6LGF2 E0VUL7 X1WK10 B4QV48 Q9VEG4 B3MTN6 Q94979 B3P0G8 A0A1B6M0P2 B4IBD8 A0A2S2PQ43 A0A2H8TYZ0 B0WKK1 B4PKI1 A0A2S2QQ37 Q8SX01 B4NFK8 U3U984 A0A336M7W2 B4JUG0 A0A1B6E925 A0A3B0K2J6 B5DV37 B5DW24 B4KDQ2 B4GMH9 A0A1A9WBU3 A0A1B0B1Q2 A0A0Q9WBH2 B4M5G8 A0A1W4W6I0 A0A2U9PG29 T1HT73 A0A1A9XYP8 A0A0R3NK35 A0A1B6FIP4 A0A1B6H2E8 A0A1B6LJ02 A0A1B6MF70 A0A1B6KV52 A0A2R7WBA7 D6WSS0 A0A0M4EIV1 A0A1B0A9S3 A0A182K8C9 A0A0K8UNB6 W5JNP0 A0A0K8VSY6 U3UA09 A0A0P6IQ05 E9HAA1 A0A0K8UBQ4 A0A034WB40 V3ZKN4 W8C5K0 A0A139WFA3 J9JW42 K1QUZ3 A0A2T7NZD4 A0A0P5NUL5 A0A2P8YYD8 A0A210PHD1
Pubmed
26354079
22118469
26227816
19121390
29403074
24845553
+ More
25244985 24438588 20966253 12364791 14747013 17210077 17510324 24466069 30735632 18362917 19820115 18054377 18025266 18316733 20068045 21843505 23604020 20566863 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8995395 17550304 23932938 15632085 20920257 23761445 21292972 25348373 23254933 24495485 22992520 28812685
25244985 24438588 20966253 12364791 14747013 17210077 17510324 24466069 30735632 18362917 19820115 18054377 18025266 18316733 20068045 21843505 23604020 20566863 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8995395 17550304 23932938 15632085 20920257 23761445 21292972 25348373 23254933 24495485 22992520 28812685
EMBL
AF177772
AAO32632.1
FJ374692
ACJ06652.1
ODYU01009094
SOQ53267.1
+ More
KQ461192 KPJ07033.1 AGBW02010040 OWR49311.1 JTDY01000855 KOB75664.1 NWSH01001389 PCG71443.1 KT031048 ALM88346.1 KQ459595 KPI95740.1 BABH01026621 CVRI01000002 CRK86947.1 PYGN01002550 PSN30300.1 KK853560 KDR06730.1 AXCM01005948 AXCM01005949 ATLV01016358 KE525079 KFB41293.1 APCN01002644 AAAB01008817 EAA05376.3 CH477302 EAT44221.2 KF711859 MH810191 AHL28506.1 QBC65456.1 NEVH01015824 PNF26617.1 KQ971352 KYB26717.1 EFA07460.2 KK853291 KDR08863.1 BK006111 DAA64492.1 MG550226 AWT50660.1 UFQS01000524 UFQT01000524 SSX04597.1 SSX24960.1 GEBQ01017184 JAT22793.1 DS235787 EEB17073.1 ABLF02040478 ABLF02040497 ABLF02040505 ABLF02040514 ABLF02050364 ABLF02058296 CM000364 EDX12531.1 AE014297 BT128700 AAF55460.2 AAN13752.1 AEF33881.1 CH902623 EDV30167.2 U47005 AH003653 AAB07030.1 AAB07999.1 CH954181 EDV48683.1 GEBQ01010468 JAT29509.1 CH480827 EDW44696.1 GGMR01018799 MBY31418.1 GFXV01007662 MBW19467.1 DS231973 EDS29895.1 CM000160 EDW95820.1 KRK02742.1 GGMS01010693 MBY79896.1 AY094926 AAM11279.1 CH964251 EDW83075.2 AB817332 BAO01099.1 SSX04596.1 SSX24959.1 CH916374 EDV91130.1 GEDC01002903 JAS34395.1 OUUW01000014 SPP88454.1 CH675291 EDY71770.2 CM000070 EDY68492.2 CH933806 EDW13886.1 CH479185 EDW38053.1 JXJN01007259 JXJN01007260 JXJN01007261 CH940652 KRF79137.1 EDW59879.2 MG550225 AWT50659.1 ACPB03006071 KRT01116.1 GECZ01019712 JAS50057.1 GECZ01000916 JAS68853.1 GEBQ01016403 JAT23574.1 GEBQ01005404 JAT34573.1 GEBQ01024664 JAT15313.1 KK854555 PTY16819.1 BK006108 DAA64489.1 CP012526 ALC46684.1 GDHF01024316 JAI27998.1 ADMH02000985 ETN64510.1 GDHF01010356 JAI41958.1 AB817331 BAO01098.1 GDIQ01001775 JAN92962.1 GL732611 EFX71271.1 GDHF01028308 JAI24006.1 GAKP01006186 JAC52766.1 KB203827 ESO82975.1 GAMC01004829 JAC01727.1 KYB26612.1 ABLF02030597 ABLF02030601 ABLF02030605 JH816438 EKC35054.1 PZQS01000008 PVD26535.1 GDIQ01137509 JAL14217.1 PYGN01000287 PSN49269.1 NEDP02076698 OWF35904.1
KQ461192 KPJ07033.1 AGBW02010040 OWR49311.1 JTDY01000855 KOB75664.1 NWSH01001389 PCG71443.1 KT031048 ALM88346.1 KQ459595 KPI95740.1 BABH01026621 CVRI01000002 CRK86947.1 PYGN01002550 PSN30300.1 KK853560 KDR06730.1 AXCM01005948 AXCM01005949 ATLV01016358 KE525079 KFB41293.1 APCN01002644 AAAB01008817 EAA05376.3 CH477302 EAT44221.2 KF711859 MH810191 AHL28506.1 QBC65456.1 NEVH01015824 PNF26617.1 KQ971352 KYB26717.1 EFA07460.2 KK853291 KDR08863.1 BK006111 DAA64492.1 MG550226 AWT50660.1 UFQS01000524 UFQT01000524 SSX04597.1 SSX24960.1 GEBQ01017184 JAT22793.1 DS235787 EEB17073.1 ABLF02040478 ABLF02040497 ABLF02040505 ABLF02040514 ABLF02050364 ABLF02058296 CM000364 EDX12531.1 AE014297 BT128700 AAF55460.2 AAN13752.1 AEF33881.1 CH902623 EDV30167.2 U47005 AH003653 AAB07030.1 AAB07999.1 CH954181 EDV48683.1 GEBQ01010468 JAT29509.1 CH480827 EDW44696.1 GGMR01018799 MBY31418.1 GFXV01007662 MBW19467.1 DS231973 EDS29895.1 CM000160 EDW95820.1 KRK02742.1 GGMS01010693 MBY79896.1 AY094926 AAM11279.1 CH964251 EDW83075.2 AB817332 BAO01099.1 SSX04596.1 SSX24959.1 CH916374 EDV91130.1 GEDC01002903 JAS34395.1 OUUW01000014 SPP88454.1 CH675291 EDY71770.2 CM000070 EDY68492.2 CH933806 EDW13886.1 CH479185 EDW38053.1 JXJN01007259 JXJN01007260 JXJN01007261 CH940652 KRF79137.1 EDW59879.2 MG550225 AWT50659.1 ACPB03006071 KRT01116.1 GECZ01019712 JAS50057.1 GECZ01000916 JAS68853.1 GEBQ01016403 JAT23574.1 GEBQ01005404 JAT34573.1 GEBQ01024664 JAT15313.1 KK854555 PTY16819.1 BK006108 DAA64489.1 CP012526 ALC46684.1 GDHF01024316 JAI27998.1 ADMH02000985 ETN64510.1 GDHF01010356 JAI41958.1 AB817331 BAO01098.1 GDIQ01001775 JAN92962.1 GL732611 EFX71271.1 GDHF01028308 JAI24006.1 GAKP01006186 JAC52766.1 KB203827 ESO82975.1 GAMC01004829 JAC01727.1 KYB26612.1 ABLF02030597 ABLF02030601 ABLF02030605 JH816438 EKC35054.1 PZQS01000008 PVD26535.1 GDIQ01137509 JAL14217.1 PYGN01000287 PSN49269.1 NEDP02076698 OWF35904.1
Proteomes
UP000053240
UP000007151
UP000037510
UP000218220
UP000053268
UP000005204
+ More
UP000183832 UP000245037 UP000027135 UP000076408 UP000075883 UP000030765 UP000075882 UP000075840 UP000007062 UP000076407 UP000075920 UP000075900 UP000008820 UP000075885 UP000075902 UP000235965 UP000007266 UP000075884 UP000075880 UP000069272 UP000192221 UP000009046 UP000007819 UP000000304 UP000000803 UP000007801 UP000008711 UP000001292 UP000002320 UP000002282 UP000007798 UP000001070 UP000268350 UP000001819 UP000009192 UP000008744 UP000091820 UP000092460 UP000008792 UP000192223 UP000015103 UP000092443 UP000092553 UP000092445 UP000075881 UP000000673 UP000000305 UP000030746 UP000005408 UP000245119 UP000242188
UP000183832 UP000245037 UP000027135 UP000076408 UP000075883 UP000030765 UP000075882 UP000075840 UP000007062 UP000076407 UP000075920 UP000075900 UP000008820 UP000075885 UP000075902 UP000235965 UP000007266 UP000075884 UP000075880 UP000069272 UP000192221 UP000009046 UP000007819 UP000000304 UP000000803 UP000007801 UP000008711 UP000001292 UP000002320 UP000002282 UP000007798 UP000001070 UP000268350 UP000001819 UP000009192 UP000008744 UP000091820 UP000092460 UP000008792 UP000192223 UP000015103 UP000092443 UP000092553 UP000092445 UP000075881 UP000000673 UP000000305 UP000030746 UP000005408 UP000245119 UP000242188
Interpro
SUPFAM
SSF57903
SSF57903
Gene 3D
ProteinModelPortal
Q86S10
B6VCU5
A0A2H1WJT0
A0A0N1PER8
A0A212F6E5
A0A0L7LJZ7
+ More
A0A2A4JJ49 A0A0S1YDB2 A0A194PX70 H9JK36 A0A1J1HGD8 A0A2P8XE68 A0A067QID7 A0A182YF55 A0A182M4B0 A0A084VTJ9 A0A182L7T4 A0A182I861 Q7PSL6 A0A182XFE1 A0A182VX05 A0A182RSK6 Q17CY4 A0A182P438 W8PGR8 A0A182TIN3 A0A2J7QDG0 A0A139WFY6 D6WU57 A0A067QJD2 A0A182NG31 A0A182J075 A0A182FA27 N1NVE8 A0A2U9PFW7 A0A336M4P4 A0A1W4V5Q2 A0A1B6LGF2 E0VUL7 X1WK10 B4QV48 Q9VEG4 B3MTN6 Q94979 B3P0G8 A0A1B6M0P2 B4IBD8 A0A2S2PQ43 A0A2H8TYZ0 B0WKK1 B4PKI1 A0A2S2QQ37 Q8SX01 B4NFK8 U3U984 A0A336M7W2 B4JUG0 A0A1B6E925 A0A3B0K2J6 B5DV37 B5DW24 B4KDQ2 B4GMH9 A0A1A9WBU3 A0A1B0B1Q2 A0A0Q9WBH2 B4M5G8 A0A1W4W6I0 A0A2U9PG29 T1HT73 A0A1A9XYP8 A0A0R3NK35 A0A1B6FIP4 A0A1B6H2E8 A0A1B6LJ02 A0A1B6MF70 A0A1B6KV52 A0A2R7WBA7 D6WSS0 A0A0M4EIV1 A0A1B0A9S3 A0A182K8C9 A0A0K8UNB6 W5JNP0 A0A0K8VSY6 U3UA09 A0A0P6IQ05 E9HAA1 A0A0K8UBQ4 A0A034WB40 V3ZKN4 W8C5K0 A0A139WFA3 J9JW42 K1QUZ3 A0A2T7NZD4 A0A0P5NUL5 A0A2P8YYD8 A0A210PHD1
A0A2A4JJ49 A0A0S1YDB2 A0A194PX70 H9JK36 A0A1J1HGD8 A0A2P8XE68 A0A067QID7 A0A182YF55 A0A182M4B0 A0A084VTJ9 A0A182L7T4 A0A182I861 Q7PSL6 A0A182XFE1 A0A182VX05 A0A182RSK6 Q17CY4 A0A182P438 W8PGR8 A0A182TIN3 A0A2J7QDG0 A0A139WFY6 D6WU57 A0A067QJD2 A0A182NG31 A0A182J075 A0A182FA27 N1NVE8 A0A2U9PFW7 A0A336M4P4 A0A1W4V5Q2 A0A1B6LGF2 E0VUL7 X1WK10 B4QV48 Q9VEG4 B3MTN6 Q94979 B3P0G8 A0A1B6M0P2 B4IBD8 A0A2S2PQ43 A0A2H8TYZ0 B0WKK1 B4PKI1 A0A2S2QQ37 Q8SX01 B4NFK8 U3U984 A0A336M7W2 B4JUG0 A0A1B6E925 A0A3B0K2J6 B5DV37 B5DW24 B4KDQ2 B4GMH9 A0A1A9WBU3 A0A1B0B1Q2 A0A0Q9WBH2 B4M5G8 A0A1W4W6I0 A0A2U9PG29 T1HT73 A0A1A9XYP8 A0A0R3NK35 A0A1B6FIP4 A0A1B6H2E8 A0A1B6LJ02 A0A1B6MF70 A0A1B6KV52 A0A2R7WBA7 D6WSS0 A0A0M4EIV1 A0A1B0A9S3 A0A182K8C9 A0A0K8UNB6 W5JNP0 A0A0K8VSY6 U3UA09 A0A0P6IQ05 E9HAA1 A0A0K8UBQ4 A0A034WB40 V3ZKN4 W8C5K0 A0A139WFA3 J9JW42 K1QUZ3 A0A2T7NZD4 A0A0P5NUL5 A0A2P8YYD8 A0A210PHD1
PDB
4AY9
E-value=9.22521e-18,
Score=223
Ontologies
KEGG
GO
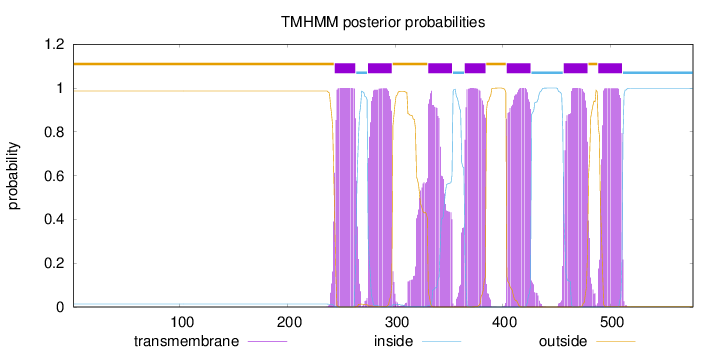
Topology
Length:
577
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
153.49735
Exp number, first 60 AAs:
0.00035
Total prob of N-in:
0.01329
outside
1 - 243
TMhelix
244 - 263
inside
264 - 274
TMhelix
275 - 297
outside
298 - 330
TMhelix
331 - 353
inside
354 - 364
TMhelix
365 - 384
outside
385 - 403
TMhelix
404 - 426
inside
427 - 456
TMhelix
457 - 479
outside
480 - 488
TMhelix
489 - 511
inside
512 - 577
Population Genetic Test Statistics
Pi
21.440348
Theta
21.708528
Tajima's D
-0.310383
CLR
1.517553
CSRT
0.287135643217839
Interpretation
Uncertain
