Pre Gene Modal
BGIBMGA009885
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_DDX47_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.185
Sequence
CDS
ATGACTGCAGACAAGGAAAGCTATGGAGACGAAACCAACCAGGATAGTGAGGTGGAACAAACTCCAACCGAAAATGTCAACGAAGATACTGAAGATGATAAAATAACATTTAAGGATTTGGGAGTTGTAGACGTACTCTGCGAGGCTTGTGAAGAATTGAAATGGAAGAAACCCTCTAAGATTCAAAAAGAAGCTATTCCCGTCGCGTTACTAGGCAAAGATATCATTGGTCTTGCCGAGACAGGTTCTGGTAAGACAGGTGCCTTTGCCCTGCCCATACTTCAAGCACTACTGGAGAACCCGCAGAGATATTTTGCACTTATACTCACACCAACAAGGGAACTGGCATTTCAGATATCAGAACAATTTGAAGCACTTGGTGCAAGCATAGGTGTAAAATGTGCAGTAATAGTTGGTGGTATGGACATGGTGGCACAAGCACTAATGCTATCCAAGAAACCTCACATAATCATCGCCACTCCTGGAAGATTGGTTGACCACCTGGAGAATACCAAGGGATTTAATTTAAGGCCTCTAAAATATTTAGTAATGGATGAAGCGGATAGAATTCTCAACATGGATTTCGAAGTGGAAGTGGACAAGATTCTCCGTGTGATACCAAGAGAACGCCACACTTATCTCTTTTCTGCAACAATGACGAAGAAAGTGCAGAAGCTCCAAAGGGCATCATTACAAGATCCTGTGAAAGTGGAGGTGTCTACAAAGTACCAGACTGTTGATAAGCTTATGCAATCCTACATCTTCATACCTGTTAAGTTTAAGGATGTATACCTAGTTCACATTCTCAACGAGATGGCTGGAAACTCGTTTATCGTCTTCGTCTCGACGTGCGCCGGTGCGCTGCGCTGTGCATTATTGCTCAGAAACTTGGGTCTGGCCGCGGTACCGCTACACGGTCAGATGTCTCAGAATAAACGTCTCGCAGCCCTGAACAAATTCAAGAGCAAATCCAGATCGGTGTTGATCTGTACTGACGTAGCATCCAGAGGTCTGGACATTCCTCACGTGGACGTCGTGATAAACCTGGACATCCCGCTGCACAGCAAGGACTACATCCACCGCGTGGGCCGCACCGCCCGCGCCGGTCGGGCCGGTAAAGCTATCACTTTTGTCACGCAGTACGACGTGGAGTTATACCAGCGAATTGAACAGTTGATCGGGAAACAGCTGCCTTTATACAAGACGGAGGAGAGCGAGGTCATGGTGCTGCAGGAGAGGGTCGCTGAGGCCCAACGTCTCACTAGAATCGAAATGAAAGAACTAGAAGACAAGAAGAGTACGAAGGGTAAGAAGCGAGGTGCCGACTCTGACGACGACACAGAGGATGCGGTCGGAGTCCGCAGACGAATCAAGGGGAAACCTAAACACGCTGGCAAGAAACGGAAACGATGA
Protein
MTADKESYGDETNQDSEVEQTPTENVNEDTEDDKITFKDLGVVDVLCEACEELKWKKPSKIQKEAIPVALLGKDIIGLAETGSGKTGAFALPILQALLENPQRYFALILTPTRELAFQISEQFEALGASIGVKCAVIVGGMDMVAQALMLSKKPHIIIATPGRLVDHLENTKGFNLRPLKYLVMDEADRILNMDFEVEVDKILRVIPRERHTYLFSATMTKKVQKLQRASLQDPVKVEVSTKYQTVDKLMQSYIFIPVKFKDVYLVHILNEMAGNSFIVFVSTCAGALRCALLLRNLGLAAVPLHGQMSQNKRLAALNKFKSKSRSVLICTDVASRGLDIPHVDVVINLDIPLHSKDYIHRVGRTARAGRAGKAITFVTQYDVELYQRIEQLIGKQLPLYKTEESEVMVLQERVAEAQRLTRIEMKELEDKKSTKGKKRGADSDDDTEDAVGVRRRIKGKPKHAGKKRKR
Summary
Similarity
Belongs to the DEAD box helicase family.
Belongs to the TUB family.
Belongs to the enoyl-CoA hydratase/isomerase family.
Belongs to the TUB family.
Belongs to the enoyl-CoA hydratase/isomerase family.
Uniprot
H9JK34
A0A212FIG4
A0A194PS03
S4NXR9
A0A2H1VSR9
A0A0N0PAJ6
+ More
A0A1L4A1S3 A0A1W4WE04 D6WVE7 B4P6Z5 A0A310SN04 A0A0C9R2J3 W8BQX6 A0A1J1HZV0 A0A1B0F9N6 A0A1A9WKS2 E0V9W5 A0A087ZQ19 A0A067QM22 A0A1I8PJ10 A0A0N0U5M2 A0A1J1HKU5 A0A1A9ZDD0 B4KGP2 A0A0Q9XIZ8 A0A154NY33 A0A1A9V9S1 A0A1B0B7P1 A0A0M4EQU0 A0A0L0C5W8 Q8MRB7 Q9VIF6 K7IXD1 A0A1A9XL24 A0A084WG48 Q29NS3 B4GJI5 A0A0L7R179 A0A232F4D8 A0A1Y1LL38 A0A182NBY8 U5EYL8 A0A2A3EHN5 B3MN83 B4JQB9 A0A1W4UI79 A0A3B0K943 B4Q3Y9 A0A1B6CR44 B4IFK8 B3NKU4 A0A1I8N8U9 J9K6I8 A0A1W4UVR9 A0A182Q3X7 Q16HQ7 A0A1S4G0Y5 J3JXB4 A0A0Q9WLS3 A0A2H8U0A3 A0A0A9X4E7 A0A1B6L8N1 A0A2M4AAB4 A0A182F1Q0 A0A2M4BL96 B4MDP0 B4N7D0 A0A034VG68 A0A182MG03 E2AHH5 A0A2M3Z4V9 A0A182KEC6 A0A1B6ILT0 A0A023ET67 A0A0T6B224 A0A182XGC0 F4W650 A0A182WGP0 T1E9Z9 A0A0A1X7Y8 A0A182RQ57 A0A2M3Z519 A0A2M4BLJ6 A0A131XM20 A0A1S4H264 A0A1B6F6D1 A0A182HRR4 Q7Q1G0 A0A182UK67 A0A2M4BLC0 A0A2M4BLB9 A0A195AVQ0 A0A182P326 A0A1S3HIV9 A0A1S3HIM4 A0A182VIW0 A0A158P1X3 W5J772 A0A182YDG7 U4TYP4
A0A1L4A1S3 A0A1W4WE04 D6WVE7 B4P6Z5 A0A310SN04 A0A0C9R2J3 W8BQX6 A0A1J1HZV0 A0A1B0F9N6 A0A1A9WKS2 E0V9W5 A0A087ZQ19 A0A067QM22 A0A1I8PJ10 A0A0N0U5M2 A0A1J1HKU5 A0A1A9ZDD0 B4KGP2 A0A0Q9XIZ8 A0A154NY33 A0A1A9V9S1 A0A1B0B7P1 A0A0M4EQU0 A0A0L0C5W8 Q8MRB7 Q9VIF6 K7IXD1 A0A1A9XL24 A0A084WG48 Q29NS3 B4GJI5 A0A0L7R179 A0A232F4D8 A0A1Y1LL38 A0A182NBY8 U5EYL8 A0A2A3EHN5 B3MN83 B4JQB9 A0A1W4UI79 A0A3B0K943 B4Q3Y9 A0A1B6CR44 B4IFK8 B3NKU4 A0A1I8N8U9 J9K6I8 A0A1W4UVR9 A0A182Q3X7 Q16HQ7 A0A1S4G0Y5 J3JXB4 A0A0Q9WLS3 A0A2H8U0A3 A0A0A9X4E7 A0A1B6L8N1 A0A2M4AAB4 A0A182F1Q0 A0A2M4BL96 B4MDP0 B4N7D0 A0A034VG68 A0A182MG03 E2AHH5 A0A2M3Z4V9 A0A182KEC6 A0A1B6ILT0 A0A023ET67 A0A0T6B224 A0A182XGC0 F4W650 A0A182WGP0 T1E9Z9 A0A0A1X7Y8 A0A182RQ57 A0A2M3Z519 A0A2M4BLJ6 A0A131XM20 A0A1S4H264 A0A1B6F6D1 A0A182HRR4 Q7Q1G0 A0A182UK67 A0A2M4BLC0 A0A2M4BLB9 A0A195AVQ0 A0A182P326 A0A1S3HIV9 A0A1S3HIM4 A0A182VIW0 A0A158P1X3 W5J772 A0A182YDG7 U4TYP4
Pubmed
19121390
22118469
26354079
23622113
18362917
19820115
+ More
17994087 17550304 24495485 20566863 24845553 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 24438588 15632085 28648823 28004739 22936249 25315136 17510324 22516182 23537049 25401762 26823975 25348373 20798317 24945155 21719571 25830018 28049606 12364791 21347285 20920257 23761445 25244985
17994087 17550304 24495485 20566863 24845553 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 24438588 15632085 28648823 28004739 22936249 25315136 17510324 22516182 23537049 25401762 26823975 25348373 20798317 24945155 21719571 25830018 28049606 12364791 21347285 20920257 23761445 25244985
EMBL
BABH01026605
BABH01026606
AGBW02008395
OWR53508.1
KQ459595
KPI95738.1
+ More
GAIX01012047 JAA80513.1 ODYU01003914 SOQ43264.1 KQ461192 KPJ07031.1 KU182489 API61864.1 KQ971357 EFA08555.1 CM000158 EDW89964.1 KQ761824 OAD56773.1 GBYB01007062 JAG76829.1 GAMC01002875 JAC03681.1 CVRI01000036 CRK93058.1 CCAG010005373 DS235000 EEB10171.1 KK853162 KDR10427.1 KQ435763 KOX75401.1 CVRI01000009 CRK88653.1 CH933807 EDW13242.1 KRG03635.1 KQ434782 KZC04527.1 JXJN01009696 CP012523 ALC39425.1 JRES01000869 KNC27642.1 AY121677 AAM52004.1 AE014134 BT133459 AAF53963.1 AFH89832.1 AAZX01000018 ATLV01023442 KE525343 KFB49192.1 CH379058 EAL34571.1 CH479184 EDW36801.1 KQ414668 KOC64597.1 NNAY01000980 OXU25631.1 GEZM01052647 JAV74362.1 GANO01001923 JAB57948.1 KZ288254 PBC30709.1 CH902620 EDV31040.1 CH916372 EDV99099.1 OUUW01000006 SPP82146.1 CM000361 CM002910 EDX05705.1 KMY91284.1 GEDC01021358 JAS15940.1 CH480833 EDW46430.1 CH954179 EDV54467.1 ABLF02037377 AXCN02000691 CH478149 EAT33783.1 APGK01035795 BT127883 KB740928 AEE62845.1 ENN77964.1 CH940661 KRF85635.1 GFXV01008158 MBW19963.1 GBHO01028730 GBHO01028729 GBRD01013550 GDHC01015560 GDHC01003395 JAG14874.1 JAG14875.1 JAG52276.1 JAQ03069.1 JAQ15234.1 GEBQ01019910 JAT20067.1 GGFK01004237 MBW37558.1 GGFJ01004699 MBW53840.1 EDW71301.1 CH964182 EDW80271.1 GAKP01018396 GAKP01018394 JAC40558.1 AXCM01008610 GL439539 EFN67130.1 GGFM01002795 MBW23546.1 GECU01019833 JAS87873.1 GAPW01001088 JAC12510.1 LJIG01016242 KRT81189.1 GL887707 EGI70185.1 GAMD01001463 JAB00128.1 GBXI01007472 JAD06820.1 GGFM01002866 MBW23617.1 GGFJ01004692 MBW53833.1 GEFH01001835 JAP66746.1 AAAB01008980 GECZ01024366 JAS45403.1 APCN01001737 EAA14161.4 GGFJ01004704 MBW53845.1 GGFJ01004691 MBW53832.1 KQ976731 KYM76318.1 ADTU01006906 ADMH02002127 ETN58634.1 KB631763 ERL85947.1
GAIX01012047 JAA80513.1 ODYU01003914 SOQ43264.1 KQ461192 KPJ07031.1 KU182489 API61864.1 KQ971357 EFA08555.1 CM000158 EDW89964.1 KQ761824 OAD56773.1 GBYB01007062 JAG76829.1 GAMC01002875 JAC03681.1 CVRI01000036 CRK93058.1 CCAG010005373 DS235000 EEB10171.1 KK853162 KDR10427.1 KQ435763 KOX75401.1 CVRI01000009 CRK88653.1 CH933807 EDW13242.1 KRG03635.1 KQ434782 KZC04527.1 JXJN01009696 CP012523 ALC39425.1 JRES01000869 KNC27642.1 AY121677 AAM52004.1 AE014134 BT133459 AAF53963.1 AFH89832.1 AAZX01000018 ATLV01023442 KE525343 KFB49192.1 CH379058 EAL34571.1 CH479184 EDW36801.1 KQ414668 KOC64597.1 NNAY01000980 OXU25631.1 GEZM01052647 JAV74362.1 GANO01001923 JAB57948.1 KZ288254 PBC30709.1 CH902620 EDV31040.1 CH916372 EDV99099.1 OUUW01000006 SPP82146.1 CM000361 CM002910 EDX05705.1 KMY91284.1 GEDC01021358 JAS15940.1 CH480833 EDW46430.1 CH954179 EDV54467.1 ABLF02037377 AXCN02000691 CH478149 EAT33783.1 APGK01035795 BT127883 KB740928 AEE62845.1 ENN77964.1 CH940661 KRF85635.1 GFXV01008158 MBW19963.1 GBHO01028730 GBHO01028729 GBRD01013550 GDHC01015560 GDHC01003395 JAG14874.1 JAG14875.1 JAG52276.1 JAQ03069.1 JAQ15234.1 GEBQ01019910 JAT20067.1 GGFK01004237 MBW37558.1 GGFJ01004699 MBW53840.1 EDW71301.1 CH964182 EDW80271.1 GAKP01018396 GAKP01018394 JAC40558.1 AXCM01008610 GL439539 EFN67130.1 GGFM01002795 MBW23546.1 GECU01019833 JAS87873.1 GAPW01001088 JAC12510.1 LJIG01016242 KRT81189.1 GL887707 EGI70185.1 GAMD01001463 JAB00128.1 GBXI01007472 JAD06820.1 GGFM01002866 MBW23617.1 GGFJ01004692 MBW53833.1 GEFH01001835 JAP66746.1 AAAB01008980 GECZ01024366 JAS45403.1 APCN01001737 EAA14161.4 GGFJ01004704 MBW53845.1 GGFJ01004691 MBW53832.1 KQ976731 KYM76318.1 ADTU01006906 ADMH02002127 ETN58634.1 KB631763 ERL85947.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000192223
UP000007266
+ More
UP000002282 UP000183832 UP000092444 UP000091820 UP000009046 UP000005203 UP000027135 UP000095300 UP000053105 UP000092445 UP000009192 UP000076502 UP000078200 UP000092460 UP000092553 UP000037069 UP000000803 UP000002358 UP000092443 UP000030765 UP000001819 UP000008744 UP000053825 UP000215335 UP000075884 UP000242457 UP000007801 UP000001070 UP000192221 UP000268350 UP000000304 UP000001292 UP000008711 UP000095301 UP000007819 UP000075886 UP000008820 UP000019118 UP000008792 UP000069272 UP000007798 UP000075883 UP000000311 UP000075881 UP000076407 UP000007755 UP000075920 UP000075900 UP000075840 UP000007062 UP000075902 UP000078540 UP000075885 UP000085678 UP000075903 UP000005205 UP000000673 UP000076408 UP000030742
UP000002282 UP000183832 UP000092444 UP000091820 UP000009046 UP000005203 UP000027135 UP000095300 UP000053105 UP000092445 UP000009192 UP000076502 UP000078200 UP000092460 UP000092553 UP000037069 UP000000803 UP000002358 UP000092443 UP000030765 UP000001819 UP000008744 UP000053825 UP000215335 UP000075884 UP000242457 UP000007801 UP000001070 UP000192221 UP000268350 UP000000304 UP000001292 UP000008711 UP000095301 UP000007819 UP000075886 UP000008820 UP000019118 UP000008792 UP000069272 UP000007798 UP000075883 UP000000311 UP000075881 UP000076407 UP000007755 UP000075920 UP000075900 UP000075840 UP000007062 UP000075902 UP000078540 UP000075885 UP000085678 UP000075903 UP000005205 UP000000673 UP000076408 UP000030742
Interpro
IPR000629
RNA-helicase_DEAD-box_CS
+ More
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR011545 DEAD/DEAH_box_helicase_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR027417 P-loop_NTPase
IPR000007 Tubby_C
IPR018376 Enoyl-CoA_hyd/isom_CS
IPR036036 SOCS_box-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR025659 Tubby-like_C
IPR029045 ClpP/crotonase-like_dom_sf
IPR001753 Enoyl-CoA_hydra/iso
IPR001496 SOCS_box
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR011545 DEAD/DEAH_box_helicase_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR027417 P-loop_NTPase
IPR000007 Tubby_C
IPR018376 Enoyl-CoA_hyd/isom_CS
IPR036036 SOCS_box-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR025659 Tubby-like_C
IPR029045 ClpP/crotonase-like_dom_sf
IPR001753 Enoyl-CoA_hydra/iso
IPR001496 SOCS_box
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JK34
A0A212FIG4
A0A194PS03
S4NXR9
A0A2H1VSR9
A0A0N0PAJ6
+ More
A0A1L4A1S3 A0A1W4WE04 D6WVE7 B4P6Z5 A0A310SN04 A0A0C9R2J3 W8BQX6 A0A1J1HZV0 A0A1B0F9N6 A0A1A9WKS2 E0V9W5 A0A087ZQ19 A0A067QM22 A0A1I8PJ10 A0A0N0U5M2 A0A1J1HKU5 A0A1A9ZDD0 B4KGP2 A0A0Q9XIZ8 A0A154NY33 A0A1A9V9S1 A0A1B0B7P1 A0A0M4EQU0 A0A0L0C5W8 Q8MRB7 Q9VIF6 K7IXD1 A0A1A9XL24 A0A084WG48 Q29NS3 B4GJI5 A0A0L7R179 A0A232F4D8 A0A1Y1LL38 A0A182NBY8 U5EYL8 A0A2A3EHN5 B3MN83 B4JQB9 A0A1W4UI79 A0A3B0K943 B4Q3Y9 A0A1B6CR44 B4IFK8 B3NKU4 A0A1I8N8U9 J9K6I8 A0A1W4UVR9 A0A182Q3X7 Q16HQ7 A0A1S4G0Y5 J3JXB4 A0A0Q9WLS3 A0A2H8U0A3 A0A0A9X4E7 A0A1B6L8N1 A0A2M4AAB4 A0A182F1Q0 A0A2M4BL96 B4MDP0 B4N7D0 A0A034VG68 A0A182MG03 E2AHH5 A0A2M3Z4V9 A0A182KEC6 A0A1B6ILT0 A0A023ET67 A0A0T6B224 A0A182XGC0 F4W650 A0A182WGP0 T1E9Z9 A0A0A1X7Y8 A0A182RQ57 A0A2M3Z519 A0A2M4BLJ6 A0A131XM20 A0A1S4H264 A0A1B6F6D1 A0A182HRR4 Q7Q1G0 A0A182UK67 A0A2M4BLC0 A0A2M4BLB9 A0A195AVQ0 A0A182P326 A0A1S3HIV9 A0A1S3HIM4 A0A182VIW0 A0A158P1X3 W5J772 A0A182YDG7 U4TYP4
A0A1L4A1S3 A0A1W4WE04 D6WVE7 B4P6Z5 A0A310SN04 A0A0C9R2J3 W8BQX6 A0A1J1HZV0 A0A1B0F9N6 A0A1A9WKS2 E0V9W5 A0A087ZQ19 A0A067QM22 A0A1I8PJ10 A0A0N0U5M2 A0A1J1HKU5 A0A1A9ZDD0 B4KGP2 A0A0Q9XIZ8 A0A154NY33 A0A1A9V9S1 A0A1B0B7P1 A0A0M4EQU0 A0A0L0C5W8 Q8MRB7 Q9VIF6 K7IXD1 A0A1A9XL24 A0A084WG48 Q29NS3 B4GJI5 A0A0L7R179 A0A232F4D8 A0A1Y1LL38 A0A182NBY8 U5EYL8 A0A2A3EHN5 B3MN83 B4JQB9 A0A1W4UI79 A0A3B0K943 B4Q3Y9 A0A1B6CR44 B4IFK8 B3NKU4 A0A1I8N8U9 J9K6I8 A0A1W4UVR9 A0A182Q3X7 Q16HQ7 A0A1S4G0Y5 J3JXB4 A0A0Q9WLS3 A0A2H8U0A3 A0A0A9X4E7 A0A1B6L8N1 A0A2M4AAB4 A0A182F1Q0 A0A2M4BL96 B4MDP0 B4N7D0 A0A034VG68 A0A182MG03 E2AHH5 A0A2M3Z4V9 A0A182KEC6 A0A1B6ILT0 A0A023ET67 A0A0T6B224 A0A182XGC0 F4W650 A0A182WGP0 T1E9Z9 A0A0A1X7Y8 A0A182RQ57 A0A2M3Z519 A0A2M4BLJ6 A0A131XM20 A0A1S4H264 A0A1B6F6D1 A0A182HRR4 Q7Q1G0 A0A182UK67 A0A2M4BLC0 A0A2M4BLB9 A0A195AVQ0 A0A182P326 A0A1S3HIV9 A0A1S3HIM4 A0A182VIW0 A0A158P1X3 W5J772 A0A182YDG7 U4TYP4
PDB
3BER
E-value=2.85113e-94,
Score=882
Ontologies
GO
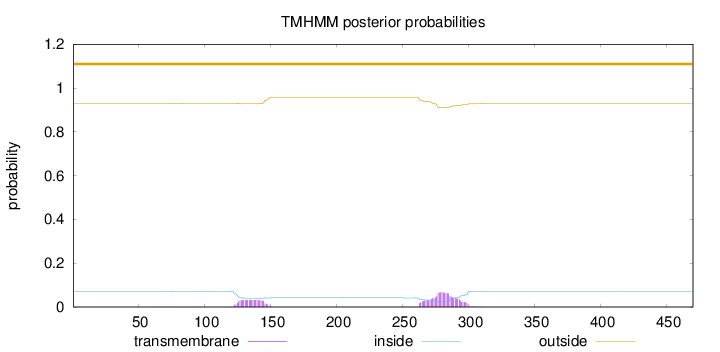
Topology
Length:
470
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.24465
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07076
outside
1 - 470
Population Genetic Test Statistics
Pi
4.962698
Theta
14.729203
Tajima's D
-1.535492
CLR
133.617464
CSRT
0.0527973601319934
Interpretation
Uncertain
