Gene
KWMTBOMO04761 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009855
Annotation
Uncharacterized_protein_OBRU01_02600_[Operophtera_brumata]
Location in the cell
Extracellular Reliability : 2.436
Sequence
CDS
ATGGCAAAGCATCATCCAGATTTAATTTTCTGCAGGAAACAGCCCGGAGTCGCTATTGGACGTTTATGTGAGAAATGTGACGGAAAATGTGTGATTTGCGATTCTTATGTGCGTCCGTGCACGTTGGTTCGTATTTGTGACGAGTGCAACTATGGCTCGTATCAGGGGCGATGTGTGATTTGCGGCGGGCCTGGAGTATCTGATGCTTATTATTGTAAGGAGTGCACGATACAGGAGAAGGATAGAGATGGCTGTCCAAAGATTGTCAATTTGGGAAGCTCTAAAACAGATCTGTTCTACGAGAGAAAGAAATATGGGTTTAGGAGACACTAG
Protein
MAKHHPDLIFCRKQPGVAIGRLCEKCDGKCVICDSYVRPCTLVRICDECNYGSYQGRCVICGGPGVSDAYYCKECTIQEKDRDGCPKIVNLGSSKTDLFYERKKYGFRRH
Summary
Uniprot
H9JK04
A0A0L7LR63
A0A1E1W4D6
A0A2H1VQZ0
A2TK64
S4PLC2
+ More
A0A0N1I6B7 A0A194PR01 A0A212FIF1 A0A182V644 A0A182KTK7 T1E830 A0A182YMS8 A0A2M4B089 A0A182F3V7 A0A182MMK7 A0A182PTD5 A0A182SMV9 A0A182NAS3 W5JW32 A0A182JTT4 A0A182UBL7 A0A084W8N1 A0A182IRA2 A0A182XJK5 Q7Q193 A0A182HRI3 A0A195EIJ6 A0A151IJW3 E9IWY4 A0A0L7QUZ6 V5FY11 F4W8Q9 A0A195B044 K7J3C4 A0A0J7KQ32 A0A067QYM8 A0A0T6B1A4 A0A195FAU3 E2B2Y8 A0A158NKF7 E2AR49 A0A2P8Z6Q5 A0A151WTA0 E0VM53 A0A026X234 A0A1B0DB07 U4TZC6 C4WXB0 A0A1Y1MS62 A0A1W4XIJ7 D7EI76 A0A1B6MRM1 W8BH77 A0A0K8U3A6 A0A034WPW3 A0A0C9RKB6 A0A232ER87 A0A3P9CTK5 A0A3P8QF90 W4ZIF2 A0A1B6MPU5 A0A1B6GGR5 A0A1B6D6S1 A0A1B6IFG0 T1HJQ1 A0A1L8DI03 A0A023EF11 A0A1Q3EZJ2 U5EZF1 B0WVS0 Q17EU3 A0A0P5H875 A0A182QTJ9 A0A0N8CV04 A0A0N7Z9F8 A0A069DNZ9 A0A1D2MCZ8 A0A0P6GKG9 A0A336LGU5 A0A226EUB7 A0A0P6FNV1 C3Z7R1 E9I4K6 A0A151NHE6 A0A0P5PVT7 A0A1B0C0F5 A0A1I8PYJ4 A0A0L0C4J0 A0A0P6EB45 A0A0P5UF27 A0A2M4B071 A0A1A9W3X7 H2Z5I2 G3NLV0 A0A0P5MAU5 A0A0P5XPB4 G1NHU8 A0A3B5KA25 A0A3Q2PEL5
A0A0N1I6B7 A0A194PR01 A0A212FIF1 A0A182V644 A0A182KTK7 T1E830 A0A182YMS8 A0A2M4B089 A0A182F3V7 A0A182MMK7 A0A182PTD5 A0A182SMV9 A0A182NAS3 W5JW32 A0A182JTT4 A0A182UBL7 A0A084W8N1 A0A182IRA2 A0A182XJK5 Q7Q193 A0A182HRI3 A0A195EIJ6 A0A151IJW3 E9IWY4 A0A0L7QUZ6 V5FY11 F4W8Q9 A0A195B044 K7J3C4 A0A0J7KQ32 A0A067QYM8 A0A0T6B1A4 A0A195FAU3 E2B2Y8 A0A158NKF7 E2AR49 A0A2P8Z6Q5 A0A151WTA0 E0VM53 A0A026X234 A0A1B0DB07 U4TZC6 C4WXB0 A0A1Y1MS62 A0A1W4XIJ7 D7EI76 A0A1B6MRM1 W8BH77 A0A0K8U3A6 A0A034WPW3 A0A0C9RKB6 A0A232ER87 A0A3P9CTK5 A0A3P8QF90 W4ZIF2 A0A1B6MPU5 A0A1B6GGR5 A0A1B6D6S1 A0A1B6IFG0 T1HJQ1 A0A1L8DI03 A0A023EF11 A0A1Q3EZJ2 U5EZF1 B0WVS0 Q17EU3 A0A0P5H875 A0A182QTJ9 A0A0N8CV04 A0A0N7Z9F8 A0A069DNZ9 A0A1D2MCZ8 A0A0P6GKG9 A0A336LGU5 A0A226EUB7 A0A0P6FNV1 C3Z7R1 E9I4K6 A0A151NHE6 A0A0P5PVT7 A0A1B0C0F5 A0A1I8PYJ4 A0A0L0C4J0 A0A0P6EB45 A0A0P5UF27 A0A2M4B071 A0A1A9W3X7 H2Z5I2 G3NLV0 A0A0P5MAU5 A0A0P5XPB4 G1NHU8 A0A3B5KA25 A0A3Q2PEL5
Pubmed
19121390
26227816
23622113
26354079
22118469
20966253
+ More
25244985 20920257 23761445 24438588 12364791 14747013 17210077 21282665 21719571 20075255 24845553 20798317 21347285 29403074 20566863 24508170 30249741 23537049 28004739 18362917 19820115 24495485 25348373 28648823 25186727 24945155 17510324 27129103 26334808 27289101 18563158 21292972 22293439 26108605 20838655 21551351
25244985 20920257 23761445 24438588 12364791 14747013 17210077 21282665 21719571 20075255 24845553 20798317 21347285 29403074 20566863 24508170 30249741 23537049 28004739 18362917 19820115 24495485 25348373 28648823 25186727 24945155 17510324 27129103 26334808 27289101 18563158 21292972 22293439 26108605 20838655 21551351
EMBL
BABH01026605
JTDY01000286
KOB77904.1
GDQN01009206
JAT81848.1
ODYU01003914
+ More
SOQ43263.1 EF210319 ABN09650.1 GAIX01004100 JAA88460.1 KQ461192 KPJ07030.1 KQ459595 KPI95737.1 AGBW02008395 OWR53509.1 GAMD01002557 JAA99033.1 GGFK01013119 MBW46440.1 AXCM01000771 ADMH02000223 GGFL01004247 ETN67325.1 MBW68425.1 ATLV01021496 KE525319 KFB46575.1 AXCP01000496 AAAB01008980 EAA13837.1 APCN01001706 KQ978881 KYN27684.1 KQ977294 KYN03940.1 GL766616 EFZ14927.1 KQ414730 KOC62445.1 GALX01005906 JAB62560.1 GL887908 EGI69550.1 KQ976692 KYM77826.1 LBMM01004498 KMQ92354.1 KK852824 KDR15457.1 LJIG01016267 KRT81113.1 KQ981693 KYN37740.1 GL445250 EFN89957.1 ADTU01018817 ADTU01018818 GL441871 EFN64088.1 PYGN01000171 PSN52183.1 KQ982762 KYQ51058.1 DS235289 EEB14459.1 KK107021 QOIP01000011 EZA62380.1 RLU16670.1 AJVK01000592 KB630546 ERL83666.1 ABLF02025841 ABLF02025843 AK342538 BAH72530.1 GEZM01025891 JAV87325.1 DS497674 EFA11678.1 GEBQ01001395 JAT38582.1 GAMC01005920 JAC00636.1 GDHF01031328 JAI20986.1 GAKP01002747 JAC56205.1 GBYB01008650 JAG78417.1 NNAY01002639 OXU20884.1 AAGJ04067662 GEBQ01001997 JAT37980.1 GECZ01008158 JAS61611.1 GEDC01015914 JAS21384.1 GECU01022069 GECU01018936 JAS85637.1 JAS88770.1 ACPB03016329 GFDF01008094 JAV05990.1 GAPW01006274 JAC07324.1 GFDL01014331 JAV20714.1 GANO01000911 JAB58960.1 DS232131 EDS35677.1 CH477279 EAT45008.1 GDIQ01233733 JAK17992.1 AXCN02000660 GDIP01096215 JAM07500.1 GDKW01000667 JAI55928.1 GBGD01003562 JAC85327.1 LJIJ01002257 LJIJ01001781 ODM89985.1 ODM90819.1 GDIQ01032344 JAN62393.1 UFQS01005502 UFQT01005502 SSX16835.1 SSX36014.1 LNIX01000002 OXA61119.1 GDIQ01062183 JAN32554.1 GG666591 EEN51416.1 GL735087 EFX61074.1 AKHW03002956 KYO36246.1 GDIQ01133073 JAL18653.1 JXJN01023588 JRES01000933 KNC27156.1 GDIQ01065286 JAN29451.1 GDIP01131062 JAL72652.1 GGFK01013120 MBW46441.1 GDIQ01158300 JAK93425.1 GDIP01069153 JAM34562.1
SOQ43263.1 EF210319 ABN09650.1 GAIX01004100 JAA88460.1 KQ461192 KPJ07030.1 KQ459595 KPI95737.1 AGBW02008395 OWR53509.1 GAMD01002557 JAA99033.1 GGFK01013119 MBW46440.1 AXCM01000771 ADMH02000223 GGFL01004247 ETN67325.1 MBW68425.1 ATLV01021496 KE525319 KFB46575.1 AXCP01000496 AAAB01008980 EAA13837.1 APCN01001706 KQ978881 KYN27684.1 KQ977294 KYN03940.1 GL766616 EFZ14927.1 KQ414730 KOC62445.1 GALX01005906 JAB62560.1 GL887908 EGI69550.1 KQ976692 KYM77826.1 LBMM01004498 KMQ92354.1 KK852824 KDR15457.1 LJIG01016267 KRT81113.1 KQ981693 KYN37740.1 GL445250 EFN89957.1 ADTU01018817 ADTU01018818 GL441871 EFN64088.1 PYGN01000171 PSN52183.1 KQ982762 KYQ51058.1 DS235289 EEB14459.1 KK107021 QOIP01000011 EZA62380.1 RLU16670.1 AJVK01000592 KB630546 ERL83666.1 ABLF02025841 ABLF02025843 AK342538 BAH72530.1 GEZM01025891 JAV87325.1 DS497674 EFA11678.1 GEBQ01001395 JAT38582.1 GAMC01005920 JAC00636.1 GDHF01031328 JAI20986.1 GAKP01002747 JAC56205.1 GBYB01008650 JAG78417.1 NNAY01002639 OXU20884.1 AAGJ04067662 GEBQ01001997 JAT37980.1 GECZ01008158 JAS61611.1 GEDC01015914 JAS21384.1 GECU01022069 GECU01018936 JAS85637.1 JAS88770.1 ACPB03016329 GFDF01008094 JAV05990.1 GAPW01006274 JAC07324.1 GFDL01014331 JAV20714.1 GANO01000911 JAB58960.1 DS232131 EDS35677.1 CH477279 EAT45008.1 GDIQ01233733 JAK17992.1 AXCN02000660 GDIP01096215 JAM07500.1 GDKW01000667 JAI55928.1 GBGD01003562 JAC85327.1 LJIJ01002257 LJIJ01001781 ODM89985.1 ODM90819.1 GDIQ01032344 JAN62393.1 UFQS01005502 UFQT01005502 SSX16835.1 SSX36014.1 LNIX01000002 OXA61119.1 GDIQ01062183 JAN32554.1 GG666591 EEN51416.1 GL735087 EFX61074.1 AKHW03002956 KYO36246.1 GDIQ01133073 JAL18653.1 JXJN01023588 JRES01000933 KNC27156.1 GDIQ01065286 JAN29451.1 GDIP01131062 JAL72652.1 GGFK01013120 MBW46441.1 GDIQ01158300 JAK93425.1 GDIP01069153 JAM34562.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000007151
UP000075903
+ More
UP000075882 UP000076408 UP000069272 UP000075883 UP000075885 UP000075901 UP000075884 UP000000673 UP000075881 UP000075902 UP000030765 UP000075880 UP000076407 UP000007062 UP000075840 UP000078492 UP000078542 UP000053825 UP000007755 UP000078540 UP000002358 UP000036403 UP000027135 UP000078541 UP000008237 UP000005205 UP000000311 UP000245037 UP000075809 UP000009046 UP000053097 UP000279307 UP000092462 UP000030742 UP000007819 UP000192223 UP000007266 UP000215335 UP000265160 UP000265100 UP000007110 UP000015103 UP000002320 UP000008820 UP000075886 UP000094527 UP000198287 UP000001554 UP000000305 UP000050525 UP000092460 UP000095300 UP000037069 UP000091820 UP000007875 UP000007635 UP000001645 UP000005226 UP000265000
UP000075882 UP000076408 UP000069272 UP000075883 UP000075885 UP000075901 UP000075884 UP000000673 UP000075881 UP000075902 UP000030765 UP000075880 UP000076407 UP000007062 UP000075840 UP000078492 UP000078542 UP000053825 UP000007755 UP000078540 UP000002358 UP000036403 UP000027135 UP000078541 UP000008237 UP000005205 UP000000311 UP000245037 UP000075809 UP000009046 UP000053097 UP000279307 UP000092462 UP000030742 UP000007819 UP000192223 UP000007266 UP000215335 UP000265160 UP000265100 UP000007110 UP000015103 UP000002320 UP000008820 UP000075886 UP000094527 UP000198287 UP000001554 UP000000305 UP000050525 UP000092460 UP000095300 UP000037069 UP000091820 UP000007875 UP000007635 UP000001645 UP000005226 UP000265000
Pfam
PF03660 PHF5
Interpro
IPR005345
PHF5
ProteinModelPortal
H9JK04
A0A0L7LR63
A0A1E1W4D6
A0A2H1VQZ0
A2TK64
S4PLC2
+ More
A0A0N1I6B7 A0A194PR01 A0A212FIF1 A0A182V644 A0A182KTK7 T1E830 A0A182YMS8 A0A2M4B089 A0A182F3V7 A0A182MMK7 A0A182PTD5 A0A182SMV9 A0A182NAS3 W5JW32 A0A182JTT4 A0A182UBL7 A0A084W8N1 A0A182IRA2 A0A182XJK5 Q7Q193 A0A182HRI3 A0A195EIJ6 A0A151IJW3 E9IWY4 A0A0L7QUZ6 V5FY11 F4W8Q9 A0A195B044 K7J3C4 A0A0J7KQ32 A0A067QYM8 A0A0T6B1A4 A0A195FAU3 E2B2Y8 A0A158NKF7 E2AR49 A0A2P8Z6Q5 A0A151WTA0 E0VM53 A0A026X234 A0A1B0DB07 U4TZC6 C4WXB0 A0A1Y1MS62 A0A1W4XIJ7 D7EI76 A0A1B6MRM1 W8BH77 A0A0K8U3A6 A0A034WPW3 A0A0C9RKB6 A0A232ER87 A0A3P9CTK5 A0A3P8QF90 W4ZIF2 A0A1B6MPU5 A0A1B6GGR5 A0A1B6D6S1 A0A1B6IFG0 T1HJQ1 A0A1L8DI03 A0A023EF11 A0A1Q3EZJ2 U5EZF1 B0WVS0 Q17EU3 A0A0P5H875 A0A182QTJ9 A0A0N8CV04 A0A0N7Z9F8 A0A069DNZ9 A0A1D2MCZ8 A0A0P6GKG9 A0A336LGU5 A0A226EUB7 A0A0P6FNV1 C3Z7R1 E9I4K6 A0A151NHE6 A0A0P5PVT7 A0A1B0C0F5 A0A1I8PYJ4 A0A0L0C4J0 A0A0P6EB45 A0A0P5UF27 A0A2M4B071 A0A1A9W3X7 H2Z5I2 G3NLV0 A0A0P5MAU5 A0A0P5XPB4 G1NHU8 A0A3B5KA25 A0A3Q2PEL5
A0A0N1I6B7 A0A194PR01 A0A212FIF1 A0A182V644 A0A182KTK7 T1E830 A0A182YMS8 A0A2M4B089 A0A182F3V7 A0A182MMK7 A0A182PTD5 A0A182SMV9 A0A182NAS3 W5JW32 A0A182JTT4 A0A182UBL7 A0A084W8N1 A0A182IRA2 A0A182XJK5 Q7Q193 A0A182HRI3 A0A195EIJ6 A0A151IJW3 E9IWY4 A0A0L7QUZ6 V5FY11 F4W8Q9 A0A195B044 K7J3C4 A0A0J7KQ32 A0A067QYM8 A0A0T6B1A4 A0A195FAU3 E2B2Y8 A0A158NKF7 E2AR49 A0A2P8Z6Q5 A0A151WTA0 E0VM53 A0A026X234 A0A1B0DB07 U4TZC6 C4WXB0 A0A1Y1MS62 A0A1W4XIJ7 D7EI76 A0A1B6MRM1 W8BH77 A0A0K8U3A6 A0A034WPW3 A0A0C9RKB6 A0A232ER87 A0A3P9CTK5 A0A3P8QF90 W4ZIF2 A0A1B6MPU5 A0A1B6GGR5 A0A1B6D6S1 A0A1B6IFG0 T1HJQ1 A0A1L8DI03 A0A023EF11 A0A1Q3EZJ2 U5EZF1 B0WVS0 Q17EU3 A0A0P5H875 A0A182QTJ9 A0A0N8CV04 A0A0N7Z9F8 A0A069DNZ9 A0A1D2MCZ8 A0A0P6GKG9 A0A336LGU5 A0A226EUB7 A0A0P6FNV1 C3Z7R1 E9I4K6 A0A151NHE6 A0A0P5PVT7 A0A1B0C0F5 A0A1I8PYJ4 A0A0L0C4J0 A0A0P6EB45 A0A0P5UF27 A0A2M4B071 A0A1A9W3X7 H2Z5I2 G3NLV0 A0A0P5MAU5 A0A0P5XPB4 G1NHU8 A0A3B5KA25 A0A3Q2PEL5
PDB
5IFE
E-value=1.04006e-49,
Score=490
Ontologies
PANTHER
Topology
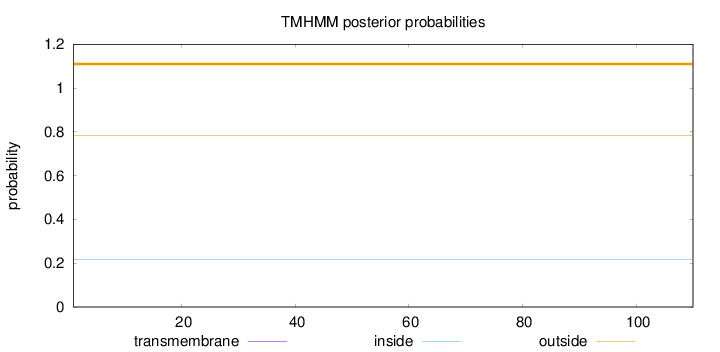
Length:
110
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00059
Exp number, first 60 AAs:
0.00025
Total prob of N-in:
0.21581
outside
1 - 110
Population Genetic Test Statistics
Pi
12.051928
Theta
21.5442
Tajima's D
-1.585083
CLR
0
CSRT
0.0496475176241188
Interpretation
Possibly Positive selection
