Gene
KWMTBOMO04760
Pre Gene Modal
BGIBMGA009884
Annotation
PREDICTED:_kinesin-like_protein_KIF12_[Bombyx_mori]
Full name
Kinesin-like protein
Location in the cell
Nuclear Reliability : 4.247
Sequence
CDS
ATGAGGAACCGCGCCGTCGGCTCGCACGCAATGAACGCTAACTCATCGCGGTCGCACACGTTGTTGTGCGTGCGCGTTCGAAGGGATCTCAGGACAGACGGTGACGTCACCTGCTCCACACACGGCAAGATCAACTTCGTCGATCTGGCCGGCAGCGAAATGACGAAACGCACTCACAGTACCGGAAAGACTTTGGAAGAGGCCAATAATATTAATAGGAGTCTCATGGTGTTAGGTTATTGCATAGCGCAACTGAGTGATGGTAAAAAAAGAGGTCATATACCATACAGGGATAGCAAGCTGACGAAGTTGTTGGCTGACAGCTTAGCCGGCAATGGGGTTACGTTAATGATAGCGTGCATTGCACCGACGCGGTCGAACTTGAACGAGACGATTAACACGCTGCGGTACGCCGCCCGAGCCAAGAAGATCAAATCTAAGCCCATCGTCAAAATGGATGCTCGTGACGCATTAATATTGAGTTTGAAACGAGAAGTGGAGGCGCTGCAAGCCGAGAACGCGCATCTGAAGACAGCGCTACACGTACAGACCGACAGCTATACTTACAGTCACAGGAACAGAACACCAAGCGATATAGAGCCGAATAAACTGTACAAGCTGCCTCAAGCCGAGCTGGTAGATCTGGTGCAGCTTCATATGGAGGAGAACACCGCGTTGAGGACGGAAAATTCTGAGCTGTTCGGCGTCAGAGACCAGCTGCTCAGGGACCAGGAGATCCTGAGCAGGGAGAACGAGAGGCTGCTGAAGAAACTGGAGGACGTTAACACAGTGTGCGTTCGTTCTCCGATTATACCAGCAAGAACGACGTACTCCAGTTCTGAGACAAACATCTGGACGAACCCGGCGACTTCCTCGGCCAACAGTGTTGCCAGCGGAGGCAGCTAA
Protein
MRNRAVGSHAMNANSSRSHTLLCVRVRRDLRTDGDVTCSTHGKINFVDLAGSEMTKRTHSTGKTLEEANNINRSLMVLGYCIAQLSDGKKRGHIPYRDSKLTKLLADSLAGNGVTLMIACIAPTRSNLNETINTLRYAARAKKIKSKPIVKMDARDALILSLKREVEALQAENAHLKTALHVQTDSYTYSHRNRTPSDIEPNKLYKLPQAELVDLVQLHMEENTALRTENSELFGVRDQLLRDQEILSRENERLLKKLEDVNTVCVRSPIIPARTTYSSSETNIWTNPATSSANSVASGGS
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the ubiquitin-activating E1 family.
Belongs to the ubiquitin-activating E1 family.
Uniprot
H9JK33
A0A212FIF9
A0A194PQK7
A0A0N1ID17
A0A336MJW8
A0A182T4M0
+ More
A0A182HFH8 Q17N88 A0A182XJW2 Q7PVB7 A0A182MQ88 A0A182U8L6 A0A2M4AIP9 B0XC45 A0A2M4AIP4 A0A1A9VQ49 A0A1B0FCR8 A0A182UPU5 A0A1S4H7S0 A0A182L508 A0A2C9GQE8 A0A2M4CP94 A0A182WQY1 A0A0L0BT97 A0A1W4VB82 A0A182QET2 A0A1A9XEY7 A0A1Y9H290 A0A1B0AD98 A0A1I8Q1R6 A0A2J7QFE5 A0A0L7RJJ0 A0A088AVS8 A0A2A3ERA4 A0A310SVC7 A0A2J7QFF4 A0A1I8MBP0 B4QB95 A0A154PNB6 A0A151WVL6 B0W2P7 A0A0N1IT26 A0A195CAM9 E2B1I6 A0A151J6L4 A0A195FKY0 A0A195BX41 A0A1Y1JW36 A0A1B0ARF4 A0A158NEC7 E2B7F8 A0A182RRY5 A0A067RFY4 F4WJ86 A0A3L8E2P7 A0A0M5IXA1 A0A3B0J5X1 A0A1W4VBP0 A0A0J9RGG5 A0A182JRA0 A1ZAV4 B3NMJ2 A0A182Y9D9 A0A126GUN4 B4P5F0 B5E083 A0A126GUN8 B4GAZ3 W8C7P5 B4MQM2 A0A182FBD8 B4J4Q2 A0A1A9W8D0 A0A1J1HJG0 B4LQ33 B4KM07 A0A1Y1JYL4 A0A0K8VRD1 A0A1Y1JYM2 A0A139WLM0 A0A1B6DPZ6 W5J7Q7 A0A1I8Q1T1 X1WTQ7 A0A026VZX5 A0A1B6CSG9 A0A1B0D8E4 A0A182IR67 A0A1D2N9S8 A0A1I8Q1T9 D2NUG1 A0A1W4UYY6 A0A226E5P7 A0A1W4VCL3 A0A0R1DSE0 A0A0J9U4P7 A0A0B4LFQ8 B3MIH9 A0A1B0C8F4
A0A182HFH8 Q17N88 A0A182XJW2 Q7PVB7 A0A182MQ88 A0A182U8L6 A0A2M4AIP9 B0XC45 A0A2M4AIP4 A0A1A9VQ49 A0A1B0FCR8 A0A182UPU5 A0A1S4H7S0 A0A182L508 A0A2C9GQE8 A0A2M4CP94 A0A182WQY1 A0A0L0BT97 A0A1W4VB82 A0A182QET2 A0A1A9XEY7 A0A1Y9H290 A0A1B0AD98 A0A1I8Q1R6 A0A2J7QFE5 A0A0L7RJJ0 A0A088AVS8 A0A2A3ERA4 A0A310SVC7 A0A2J7QFF4 A0A1I8MBP0 B4QB95 A0A154PNB6 A0A151WVL6 B0W2P7 A0A0N1IT26 A0A195CAM9 E2B1I6 A0A151J6L4 A0A195FKY0 A0A195BX41 A0A1Y1JW36 A0A1B0ARF4 A0A158NEC7 E2B7F8 A0A182RRY5 A0A067RFY4 F4WJ86 A0A3L8E2P7 A0A0M5IXA1 A0A3B0J5X1 A0A1W4VBP0 A0A0J9RGG5 A0A182JRA0 A1ZAV4 B3NMJ2 A0A182Y9D9 A0A126GUN4 B4P5F0 B5E083 A0A126GUN8 B4GAZ3 W8C7P5 B4MQM2 A0A182FBD8 B4J4Q2 A0A1A9W8D0 A0A1J1HJG0 B4LQ33 B4KM07 A0A1Y1JYL4 A0A0K8VRD1 A0A1Y1JYM2 A0A139WLM0 A0A1B6DPZ6 W5J7Q7 A0A1I8Q1T1 X1WTQ7 A0A026VZX5 A0A1B6CSG9 A0A1B0D8E4 A0A182IR67 A0A1D2N9S8 A0A1I8Q1T9 D2NUG1 A0A1W4UYY6 A0A226E5P7 A0A1W4VCL3 A0A0R1DSE0 A0A0J9U4P7 A0A0B4LFQ8 B3MIH9 A0A1B0C8F4
Pubmed
19121390
22118469
26354079
26483478
17510324
12364791
+ More
20966253 26108605 25315136 17994087 20798317 28004739 21347285 24845553 21719571 30249741 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25244985 17550304 15632085 24495485 18362917 19820115 20920257 23761445 24508170 27289101 26109357 26109356
20966253 26108605 25315136 17994087 20798317 28004739 21347285 24845553 21719571 30249741 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25244985 17550304 15632085 24495485 18362917 19820115 20920257 23761445 24508170 27289101 26109357 26109356
EMBL
BABH01026604
AGBW02008395
OWR53510.1
KQ459595
KPI95736.1
KQ461192
+ More
KPJ07029.1 UFQT01001369 SSX30230.1 JXUM01134309 KQ568101 KXJ69141.1 CH477201 EAT48166.1 AAAB01008986 EAA00098.5 AXCM01003287 GGFK01007330 MBW40651.1 DS232674 EDS44651.1 GGFK01007344 MBW40665.1 CCAG010006642 APCN01002415 GGFL01002966 MBW67144.1 JRES01001376 KNC23272.1 AXCN02001090 NEVH01015298 PNF27307.1 KQ414582 KOC70891.1 KZ288198 PBC33659.1 KQ759817 OAD62769.1 PNF27308.1 CM000362 EDX07519.1 KQ434998 KZC13333.1 KQ982696 KYQ51979.1 DS231828 EDS29800.1 KQ435922 KOX68474.1 KQ978023 KYM97872.1 GL444934 EFN60443.1 KQ979851 KYN18845.1 KQ981490 KYN41073.1 KQ976401 KYM92526.1 GEZM01099410 JAV53333.1 JXJN01002382 ADTU01013250 ADTU01013251 GL446181 EFN88333.1 KK852529 KDR21963.1 GL888182 EGI65664.1 QOIP01000001 RLU26469.1 CP012524 ALC41048.1 OUUW01000001 SPP75263.1 CM002911 KMY94619.1 AE013599 AAF57836.2 CH954179 EDV54931.2 ALI30183.1 CM000158 EDW91781.2 CM000071 EDY68943.1 ALI30184.1 CH479181 EDW32095.1 GAMC01000407 JAC06149.1 CH963849 EDW74411.1 CH916367 EDW00598.1 CVRI01000006 CRK88043.1 CH940648 EDW60356.1 CH933808 EDW10796.2 GEZM01099415 JAV53328.1 GDHF01010921 JAI41393.1 GEZM01099412 JAV53331.1 KQ971319 KYB28909.1 GEDC01009615 JAS27683.1 ADMH02002004 ETN60011.1 ABLF02023429 ABLF02023430 KK107522 EZA49317.1 GEDC01020809 JAS16489.1 AJVK01027381 LJIJ01000129 ODN02010.1 BT120104 ADB11089.1 AFH08151.1 LNIX01000006 OXA52630.1 KRK00073.1 KMY94620.1 AHN56335.1 CH902619 EDV37027.2 AJWK01000536 AJWK01000537
KPJ07029.1 UFQT01001369 SSX30230.1 JXUM01134309 KQ568101 KXJ69141.1 CH477201 EAT48166.1 AAAB01008986 EAA00098.5 AXCM01003287 GGFK01007330 MBW40651.1 DS232674 EDS44651.1 GGFK01007344 MBW40665.1 CCAG010006642 APCN01002415 GGFL01002966 MBW67144.1 JRES01001376 KNC23272.1 AXCN02001090 NEVH01015298 PNF27307.1 KQ414582 KOC70891.1 KZ288198 PBC33659.1 KQ759817 OAD62769.1 PNF27308.1 CM000362 EDX07519.1 KQ434998 KZC13333.1 KQ982696 KYQ51979.1 DS231828 EDS29800.1 KQ435922 KOX68474.1 KQ978023 KYM97872.1 GL444934 EFN60443.1 KQ979851 KYN18845.1 KQ981490 KYN41073.1 KQ976401 KYM92526.1 GEZM01099410 JAV53333.1 JXJN01002382 ADTU01013250 ADTU01013251 GL446181 EFN88333.1 KK852529 KDR21963.1 GL888182 EGI65664.1 QOIP01000001 RLU26469.1 CP012524 ALC41048.1 OUUW01000001 SPP75263.1 CM002911 KMY94619.1 AE013599 AAF57836.2 CH954179 EDV54931.2 ALI30183.1 CM000158 EDW91781.2 CM000071 EDY68943.1 ALI30184.1 CH479181 EDW32095.1 GAMC01000407 JAC06149.1 CH963849 EDW74411.1 CH916367 EDW00598.1 CVRI01000006 CRK88043.1 CH940648 EDW60356.1 CH933808 EDW10796.2 GEZM01099415 JAV53328.1 GDHF01010921 JAI41393.1 GEZM01099412 JAV53331.1 KQ971319 KYB28909.1 GEDC01009615 JAS27683.1 ADMH02002004 ETN60011.1 ABLF02023429 ABLF02023430 KK107522 EZA49317.1 GEDC01020809 JAS16489.1 AJVK01027381 LJIJ01000129 ODN02010.1 BT120104 ADB11089.1 AFH08151.1 LNIX01000006 OXA52630.1 KRK00073.1 KMY94620.1 AHN56335.1 CH902619 EDV37027.2 AJWK01000536 AJWK01000537
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000075901
UP000069940
+ More
UP000249989 UP000008820 UP000076407 UP000007062 UP000075883 UP000075902 UP000002320 UP000078200 UP000092444 UP000075903 UP000075882 UP000075840 UP000075920 UP000037069 UP000192221 UP000075886 UP000092443 UP000075884 UP000092445 UP000095300 UP000235965 UP000053825 UP000005203 UP000242457 UP000095301 UP000000304 UP000076502 UP000075809 UP000053105 UP000078542 UP000000311 UP000078492 UP000078541 UP000078540 UP000092460 UP000005205 UP000008237 UP000075900 UP000027135 UP000007755 UP000279307 UP000092553 UP000268350 UP000075881 UP000000803 UP000008711 UP000076408 UP000002282 UP000001819 UP000008744 UP000007798 UP000069272 UP000001070 UP000091820 UP000183832 UP000008792 UP000009192 UP000007266 UP000000673 UP000007819 UP000053097 UP000092462 UP000075880 UP000094527 UP000198287 UP000007801 UP000092461
UP000249989 UP000008820 UP000076407 UP000007062 UP000075883 UP000075902 UP000002320 UP000078200 UP000092444 UP000075903 UP000075882 UP000075840 UP000075920 UP000037069 UP000192221 UP000075886 UP000092443 UP000075884 UP000092445 UP000095300 UP000235965 UP000053825 UP000005203 UP000242457 UP000095301 UP000000304 UP000076502 UP000075809 UP000053105 UP000078542 UP000000311 UP000078492 UP000078541 UP000078540 UP000092460 UP000005205 UP000008237 UP000075900 UP000027135 UP000007755 UP000279307 UP000092553 UP000268350 UP000075881 UP000000803 UP000008711 UP000076408 UP000002282 UP000001819 UP000008744 UP000007798 UP000069272 UP000001070 UP000091820 UP000183832 UP000008792 UP000009192 UP000007266 UP000000673 UP000007819 UP000053097 UP000092462 UP000075880 UP000094527 UP000198287 UP000007801 UP000092461
Pfam
Interpro
IPR028763
Kif12
+ More
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR001752 Kinesin_motor_dom
IPR027640 Kinesin-like_fam
IPR036961 Kinesin_motor_dom_sf
IPR036284 GGL_sf
IPR015898 G-protein_gamma-like_dom
IPR038441 THAP_Znf_sf
IPR006612 THAP_Znf
IPR032418 E1_FCCH
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR042302 E1_FCCH_sf
IPR042063 Ubi_acti_E1_SCCH
IPR032420 E1_4HB
IPR018075 UBQ-activ_enz_E1
IPR035985 Ubiquitin-activating_enz
IPR038252 UBA_E1_C_sf
IPR000594 ThiF_NAD_FAD-bd
IPR019572 UBA_E1_Cys
IPR018965 Ub-activating_enz_E1_C
IPR000011 UBQ/SUMO-activ_enz_E1-like
IPR019787 Znf_PHD-finger
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR011011 Znf_FYVE_PHD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR011990 TPR-like_helical_dom_sf
IPR039941 TT30
IPR019821 Kinesin_motor_CS
IPR027417 P-loop_NTPase
IPR001752 Kinesin_motor_dom
IPR027640 Kinesin-like_fam
IPR036961 Kinesin_motor_dom_sf
IPR036284 GGL_sf
IPR015898 G-protein_gamma-like_dom
IPR038441 THAP_Znf_sf
IPR006612 THAP_Znf
IPR032418 E1_FCCH
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR042302 E1_FCCH_sf
IPR042063 Ubi_acti_E1_SCCH
IPR032420 E1_4HB
IPR018075 UBQ-activ_enz_E1
IPR035985 Ubiquitin-activating_enz
IPR038252 UBA_E1_C_sf
IPR000594 ThiF_NAD_FAD-bd
IPR019572 UBA_E1_Cys
IPR018965 Ub-activating_enz_E1_C
IPR000011 UBQ/SUMO-activ_enz_E1-like
IPR019787 Znf_PHD-finger
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR011011 Znf_FYVE_PHD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR011990 TPR-like_helical_dom_sf
IPR039941 TT30
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JK33
A0A212FIF9
A0A194PQK7
A0A0N1ID17
A0A336MJW8
A0A182T4M0
+ More
A0A182HFH8 Q17N88 A0A182XJW2 Q7PVB7 A0A182MQ88 A0A182U8L6 A0A2M4AIP9 B0XC45 A0A2M4AIP4 A0A1A9VQ49 A0A1B0FCR8 A0A182UPU5 A0A1S4H7S0 A0A182L508 A0A2C9GQE8 A0A2M4CP94 A0A182WQY1 A0A0L0BT97 A0A1W4VB82 A0A182QET2 A0A1A9XEY7 A0A1Y9H290 A0A1B0AD98 A0A1I8Q1R6 A0A2J7QFE5 A0A0L7RJJ0 A0A088AVS8 A0A2A3ERA4 A0A310SVC7 A0A2J7QFF4 A0A1I8MBP0 B4QB95 A0A154PNB6 A0A151WVL6 B0W2P7 A0A0N1IT26 A0A195CAM9 E2B1I6 A0A151J6L4 A0A195FKY0 A0A195BX41 A0A1Y1JW36 A0A1B0ARF4 A0A158NEC7 E2B7F8 A0A182RRY5 A0A067RFY4 F4WJ86 A0A3L8E2P7 A0A0M5IXA1 A0A3B0J5X1 A0A1W4VBP0 A0A0J9RGG5 A0A182JRA0 A1ZAV4 B3NMJ2 A0A182Y9D9 A0A126GUN4 B4P5F0 B5E083 A0A126GUN8 B4GAZ3 W8C7P5 B4MQM2 A0A182FBD8 B4J4Q2 A0A1A9W8D0 A0A1J1HJG0 B4LQ33 B4KM07 A0A1Y1JYL4 A0A0K8VRD1 A0A1Y1JYM2 A0A139WLM0 A0A1B6DPZ6 W5J7Q7 A0A1I8Q1T1 X1WTQ7 A0A026VZX5 A0A1B6CSG9 A0A1B0D8E4 A0A182IR67 A0A1D2N9S8 A0A1I8Q1T9 D2NUG1 A0A1W4UYY6 A0A226E5P7 A0A1W4VCL3 A0A0R1DSE0 A0A0J9U4P7 A0A0B4LFQ8 B3MIH9 A0A1B0C8F4
A0A182HFH8 Q17N88 A0A182XJW2 Q7PVB7 A0A182MQ88 A0A182U8L6 A0A2M4AIP9 B0XC45 A0A2M4AIP4 A0A1A9VQ49 A0A1B0FCR8 A0A182UPU5 A0A1S4H7S0 A0A182L508 A0A2C9GQE8 A0A2M4CP94 A0A182WQY1 A0A0L0BT97 A0A1W4VB82 A0A182QET2 A0A1A9XEY7 A0A1Y9H290 A0A1B0AD98 A0A1I8Q1R6 A0A2J7QFE5 A0A0L7RJJ0 A0A088AVS8 A0A2A3ERA4 A0A310SVC7 A0A2J7QFF4 A0A1I8MBP0 B4QB95 A0A154PNB6 A0A151WVL6 B0W2P7 A0A0N1IT26 A0A195CAM9 E2B1I6 A0A151J6L4 A0A195FKY0 A0A195BX41 A0A1Y1JW36 A0A1B0ARF4 A0A158NEC7 E2B7F8 A0A182RRY5 A0A067RFY4 F4WJ86 A0A3L8E2P7 A0A0M5IXA1 A0A3B0J5X1 A0A1W4VBP0 A0A0J9RGG5 A0A182JRA0 A1ZAV4 B3NMJ2 A0A182Y9D9 A0A126GUN4 B4P5F0 B5E083 A0A126GUN8 B4GAZ3 W8C7P5 B4MQM2 A0A182FBD8 B4J4Q2 A0A1A9W8D0 A0A1J1HJG0 B4LQ33 B4KM07 A0A1Y1JYL4 A0A0K8VRD1 A0A1Y1JYM2 A0A139WLM0 A0A1B6DPZ6 W5J7Q7 A0A1I8Q1T1 X1WTQ7 A0A026VZX5 A0A1B6CSG9 A0A1B0D8E4 A0A182IR67 A0A1D2N9S8 A0A1I8Q1T9 D2NUG1 A0A1W4UYY6 A0A226E5P7 A0A1W4VCL3 A0A0R1DSE0 A0A0J9U4P7 A0A0B4LFQ8 B3MIH9 A0A1B0C8F4
PDB
3ZFD
E-value=3.13465e-36,
Score=379
Ontologies
GO
PANTHER
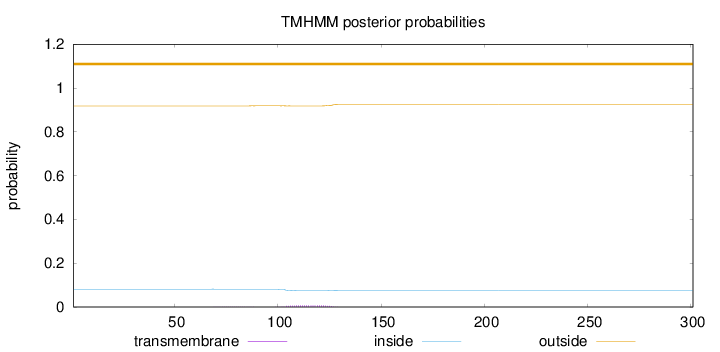
Topology
Length:
301
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17737
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08159
outside
1 - 301
Population Genetic Test Statistics
Pi
4.928917
Theta
13.458309
Tajima's D
-1.635453
CLR
225.254919
CSRT
0.0420978951052447
Interpretation
Uncertain
