Gene
KWMTBOMO04757 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009857
Annotation
PREDICTED:_myosin-IB_isoform_X1_[Papilio_xuthus]
Full name
Unconventional myosin IC
Alternative Name
Brush border myosin IB
Myosin-IB
Unconventional myosin 1C
Myosin-IB
Unconventional myosin 1C
Location in the cell
Nuclear Reliability : 2.688
Sequence
CDS
ATGGAGCACTCCCTGCAGCATCGCGAGCGGGTCGGAGTCCAAGACTTCGTTTTGCTGGAGGACTACCGATCCGAAGCGGCCTTCATTGACAATTTGAAGAAACGTTTCCATGAAAACATTATTTATACTTACATCGGTAATGTACTGATCTCGGTGAATCCGTACAAGAATCTACCGATTTACACTGAAGAGAAAACCAAACTTTACTTCAAGAAAGCATTCTTTGAGGCACCGCCGCACGTTTTCGCGATCGCCGACAACGCTTATCGGTCTTTGGTTTACGAGCACAGAGAGCAATGTATTTTGATCTCAGGCGAATCCGGCTCGGGCAAAACGGAAGCGTCGAAGAAAGTTCTGGAATACATAGCCGCCAGAACGAACCACCTCCGGAACGTGGAAAACGTGAAAGATAAGCTGTTACAAAGCAACCCGCTGCTCGAAGCGTTCGGTAACGCGAAGACGCACCGCAACGACAATTCCAGTCGCTTCGGCAAATACATGGACATCCAGTTCAACTACGAAGGCGGGCCCGAAGGTGGTCACATCCTGAACTATCTGCTGGAAAAGTCCAGGGTCGTCAGTCAGATGCATGGCGAGAGGAACTTCCACATATTCTATCAGCTACTAGCTAGCAGCGATCAGTCACTGATGACTCATTTGAAGCTTCAAGGAAGACCTGAAGCTTATAAGTACACTTCCGATTCGACCTCGCACATGAGTCAGAGAGCGAACGACCAGGAACAGTTCCGTGTCGTACAAGAGGCCATGAAGGTCATAGAAATCGGCGAGAGTGAACAGCGGGAGATCTTTGAGATCGTGGCCAGCGTCCTGCACCTCGGCAACGTGAAGTTCGTCCAGAATGACAAGGGCTATGCCGAGATCCTGTCCCACGACGCCAACAGTGGGAATGCTGCTGATCTCCTAAAAGTGAACGCGACGGCATTACGCGAGGCCCTGACCAATAGGACGATAGAGGCCCGCGGCGACGTCGTCAGCACACCCCTGGACGTGGAACAGGCTCAATACGCTAGAGACGCTCTGGCTAAAGCTATATACGACAAGCACTTCAGCTGGCTGGTCTCCAGGCTAAACTCGTCGCTTGCGCCCATCGAGAAGGACGCTAAATCTTCTGTTATAGGAATCTTGGATATCTACGGCTTTGAGATATTCCCTAAGAACAGCTTCGAGCAGTTCTGCATAAACTTCTGTAACGAGAAGCTCCAGCAGCTGTTCATCCAGCTCACGCTGCGGCAGGAGCAGGAGGAGTACCTGCGGGAAGGCATCGAGTGGGAGCCGGTCGAGTACTTCAACAATATCATCATTTGCGACCTTATCGAAGCCAGGCATAAGGGTATAATATCAATACTGGACGACGAGTGCCTTCGTCCCGGGGACGCGACAGACGCGAGTTTCCTCGACAAGCTGAACCAGCATCTGGACGGCCACCAGCACTACAAGTCCCACCGCAAATCCGACACCAAGACCCAGAAGCTGATGGGCCGTGATGAGTTCTGCCTGGTCCACTACGCCGGCGAAGTGACTTACAACGTGAACACCTTCTTGGAGAAGAACAACGATCTCCTCTTCCGCGACATCCAGAGCCTGATGGCGTCCAGCGACAACACCATCGTCGGCTGCTGCTTCAAGGACTACAACTTGAGGAGCAAGAAACGTCCGGAGACCGCCATCACTCAGTTCAAGAATTCTCTGAACGACCTCATCAAGATCCTCAGCAGCAAGGAGCCCTCGTACATCCGCTGCATCAAGCCCAACGACTTCAAGGCGCCGATGCAATTCGACGACAAGCTGGTCTCCCACCAGGTGAAGTATCTCGGTCTGATGGAGAACCTCAGGGTGCGGCGCGCCGGCTTCGCTTACAGGAGGACTTACGAAGCGTTCTTGGAAAGGTACAAGTGCCTGAGCGCGGAGACGTGGCCCAACTACCGCGGCGCCGCGCGGGACGGCGTGCAGCGGCTGGTGGAGGCCCTCCAGTACGAGAAAGAGGAATACAGGATGGGGAATACGAAAATCTTCGTTCGATTCCCGAAGACGCTCTTCGCAACGGAAGACGCCTTCCAGATAAAGAAGAACGACATAGCCACCATCATCCAGAGCAGGTGGCGAGGTTACTACCTCAGGAAGAGGTACCTCCGAATGCGGAACGCGGCCATCGTCATCCAGAAGTGGGTGAGGAGGTTCCTGGCCCAAAGACTCAGGGAGAGGAGGAGGAAAGCCGCTGACGTCATCAGAGCTTTTATTAAAGGCTTCATCACTCGCAACGGTCCGGAGACGCCAGAAAACCGGCGATTCCTCGGCGTCGCCAAGGTGCACTGGCTGAAGCGTCTCTCGGCGCAGCTCCCAACCAAGCTGCTGGACCTGTCGTGGCCGCCGTGCCCCTCCACCTGCCGAGAGGCCTCCGAGGAGCTCCATCGCCTGCACCGGGCCCACCTCGCCAGGAAGTACAGGCTGGCCTTGTCTCCTGACGATAAGAAGCAGTTTGAGCTGAAGGTCTTGGCCGAAAAAATATTTAAGGGTAAAAAGAACAGCTACCTCAGCAGCGTCGGGGAGAGGTTCGTGAACGACCGCATGAGTCCCGAGCACAGGACCTTGAGGGACACCTTCATGGCGTCGCCGGCCTGGCCGAGCGGCGAGGAACTCATCTACTCGTGCGAGGCGGTGAAGTACGACCGGCGCGGCTACAAGGCCCGCGCGCGCGGCCTGCTGGCGTCCCGCGCGGCGCTGTACGTGCTGGACGCGGGCGGGCGCCGCACGTTCCGGCTCAAGCACCGGCTGCCGCTGGACCGCCTCACCGTCGTCGTCACCAACGAGTCCGACTCGCTGCTGCTGGTCAAGGTGCCGCGCGACCTCAAGAAGGACAAGGGAGATCTGATCATATCCGTGACGCATCTCATCGAGGCCCTGACCATCGTGACGGACTACACCAAGAAGCCGGAGCTCATTGAGATAGTCGACACCAGGACCATCGCGCACTCCCTGGTCAACGGCAAGCAGGGGGGTACCATCGAGGTGACGAAGGGCACCCAGCCCGCCATACAACGCGCCAAGAGCGGGAACCTGCTCGTGGTTGCGACTCCTTAG
Protein
MEHSLQHRERVGVQDFVLLEDYRSEAAFIDNLKKRFHENIIYTYIGNVLISVNPYKNLPIYTEEKTKLYFKKAFFEAPPHVFAIADNAYRSLVYEHREQCILISGESGSGKTEASKKVLEYIAARTNHLRNVENVKDKLLQSNPLLEAFGNAKTHRNDNSSRFGKYMDIQFNYEGGPEGGHILNYLLEKSRVVSQMHGERNFHIFYQLLASSDQSLMTHLKLQGRPEAYKYTSDSTSHMSQRANDQEQFRVVQEAMKVIEIGESEQREIFEIVASVLHLGNVKFVQNDKGYAEILSHDANSGNAADLLKVNATALREALTNRTIEARGDVVSTPLDVEQAQYARDALAKAIYDKHFSWLVSRLNSSLAPIEKDAKSSVIGILDIYGFEIFPKNSFEQFCINFCNEKLQQLFIQLTLRQEQEEYLREGIEWEPVEYFNNIIICDLIEARHKGIISILDDECLRPGDATDASFLDKLNQHLDGHQHYKSHRKSDTKTQKLMGRDEFCLVHYAGEVTYNVNTFLEKNNDLLFRDIQSLMASSDNTIVGCCFKDYNLRSKKRPETAITQFKNSLNDLIKILSSKEPSYIRCIKPNDFKAPMQFDDKLVSHQVKYLGLMENLRVRRAGFAYRRTYEAFLERYKCLSAETWPNYRGAARDGVQRLVEALQYEKEEYRMGNTKIFVRFPKTLFATEDAFQIKKNDIATIIQSRWRGYYLRKRYLRMRNAAIVIQKWVRRFLAQRLRERRRKAADVIRAFIKGFITRNGPETPENRRFLGVAKVHWLKRLSAQLPTKLLDLSWPPCPSTCREASEELHRLHRAHLARKYRLALSPDDKKQFELKVLAEKIFKGKKNSYLSSVGERFVNDRMSPEHRTLRDTFMASPAWPSGEELIYSCEAVKYDRRGYKARARGLLASRAALYVLDAGGRRTFRLKHRLPLDRLTVVVTNESDSLLLVKVPRDLKKDKGDLIISVTHLIEALTIVTDYTKKPELIEIVDTRTIAHSLVNGKQGGTIEVTKGTQPAIQRAKSGNLLVVATP
Summary
Description
Unconventional myosin that functions as actin-based motor protein with ATPase activity (PubMed:30467170). Binds to membranes enriched in phosphatidylinositol 4-5-bisphosphate, and can glide along actin filaments when anchored to a lipid bilayer (PubMed:30467170). Functions as antagonist for Myo31DF, an unconventional myosin with an essential role in the establishment of body left-right asymmetry (PubMed:16598258, PubMed:18521948, PubMed:22491943, PubMed:25659376, PubMed:30467170).
Subunit
Binds F-actin.
Miscellaneous
Overexpression in larval epidermis causes sinistral twisting of the whole larval body. Ectopic expression in tracheal precursor cells causes sinistral spiraling of tracheal branches, giving rise to a spiraling ribbon shape instead of the normal smooth tube. Ectopic expression in epithelial cells causes increased elongation and a clear shift of the membrane orientation toward one side, so that the membrane is no longer perpendicular to the anterior-posterior axis (PubMed:30467170). Overexpression causes reversal of the normal left-right laterality of the embryonic midgut and hindgut (PubMed:16598258, PubMed:18521948). Overexpression causes loss of the normal dextral rotation of the male genitalia, and leads to sinistral rotation in some cases (PubMed:22491943).
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Keywords
Actin-binding
Alternative splicing
ATP-binding
Calmodulin-binding
Cell membrane
Complete proteome
Cytoplasm
Lipid-binding
Membrane
Motor protein
Myosin
Nucleotide-binding
Phosphoprotein
Reference proteome
Repeat
Feature
chain Unconventional myosin IC
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A194PRZ8
A0A0N1INH8
A0A1E1WK14
A0A1E1W8K0
A0A2A4JZ64
A0A3S2MAG9
+ More
H9JK06 A0A0L7L631 A0A348G646 A0A151J6J8 E2B7F9 A0A0C9QXW7 A0A026VZT4 A0A195CCQ9 A0A088AVI4 A0A158NEW1 A0A151WVP6 A0A195BVC0 A0A310ST08 A0A1L8E5X6 A0A182FKP9 A0A2J7QFE4 A0A2J7QFE9 F4WJ87 A0A2M4AA58 U5EW26 A0A0Q9XL00 B4L0P1 A0A2M4A9P6 W5JML8 A0A182QYK2 A0A1Q3FLX3 A0A1Q3FLL4 A0A1A9V471 A0A182YL50 A0A1A9YEG4 A0A1B0B7X8 A0A1Q3FLT0 A0A1Q3FLT2 A0A1Q3FM21 A0A0M3QVU1 B4IXH1 A0A0Q9WHM7 A0A182H660 B4LG69 A0A182KAK6 A0A1S4FUJ9 Q16NP7 B0X747 A0A0M8ZS21 A0A182TTN5 A0A182VJX7 A0A182L4J8 A0A1A9ZS32 A0A182IYV6 A0A182WED5 A0A0P8ZU70 A0A084VBX9 B3M521 A0A1A9WLI6 A0A3B0KL26 A0A182PEJ1 Q29EW2 A0A0R3P5Y2 B4N3T9 A0A1L8EGC1 T1P833 B4PD73 A0A0R1DZQ6 A0A1I8N9X0 A0A1I8N9W3 A0A2A3EPN8 H8F4R0 A0A0L0CIE1 Q23979 Q23979-2 Q23979-3 A0A0Q5UH73 B3NB94 A0A0Q5U5L1 A0A0A1WZ38 A0A0A1WHX9 A0A034W3K4 A0A034W101 A0A0J9RLM0 W8B7N0 A0A0J9RLH4 A0A0K8V3T3 A0A0K8UZK9 A0A0J9RN04 A0A1W4W7N5 B4HVR8 A0A1W4VV78 A0A1W4W748 A0A1I8P1M8 A0A182R4F7 A0A182HH34 A0A023FBM3 A0A1B6CDD8 A0A1Y1JZT2
H9JK06 A0A0L7L631 A0A348G646 A0A151J6J8 E2B7F9 A0A0C9QXW7 A0A026VZT4 A0A195CCQ9 A0A088AVI4 A0A158NEW1 A0A151WVP6 A0A195BVC0 A0A310ST08 A0A1L8E5X6 A0A182FKP9 A0A2J7QFE4 A0A2J7QFE9 F4WJ87 A0A2M4AA58 U5EW26 A0A0Q9XL00 B4L0P1 A0A2M4A9P6 W5JML8 A0A182QYK2 A0A1Q3FLX3 A0A1Q3FLL4 A0A1A9V471 A0A182YL50 A0A1A9YEG4 A0A1B0B7X8 A0A1Q3FLT0 A0A1Q3FLT2 A0A1Q3FM21 A0A0M3QVU1 B4IXH1 A0A0Q9WHM7 A0A182H660 B4LG69 A0A182KAK6 A0A1S4FUJ9 Q16NP7 B0X747 A0A0M8ZS21 A0A182TTN5 A0A182VJX7 A0A182L4J8 A0A1A9ZS32 A0A182IYV6 A0A182WED5 A0A0P8ZU70 A0A084VBX9 B3M521 A0A1A9WLI6 A0A3B0KL26 A0A182PEJ1 Q29EW2 A0A0R3P5Y2 B4N3T9 A0A1L8EGC1 T1P833 B4PD73 A0A0R1DZQ6 A0A1I8N9X0 A0A1I8N9W3 A0A2A3EPN8 H8F4R0 A0A0L0CIE1 Q23979 Q23979-2 Q23979-3 A0A0Q5UH73 B3NB94 A0A0Q5U5L1 A0A0A1WZ38 A0A0A1WHX9 A0A034W3K4 A0A034W101 A0A0J9RLM0 W8B7N0 A0A0J9RLH4 A0A0K8V3T3 A0A0K8UZK9 A0A0J9RN04 A0A1W4W7N5 B4HVR8 A0A1W4VV78 A0A1W4W748 A0A1I8P1M8 A0A182R4F7 A0A182HH34 A0A023FBM3 A0A1B6CDD8 A0A1Y1JZT2
Pubmed
26354079
19121390
26227816
20798317
24508170
21347285
+ More
21719571 17994087 18057021 20920257 23761445 25244985 26483478 17510324 20966253 24438588 15632085 23185243 17550304 25315136 26108605 8201616 10731132 12537572 12537569 8216259 7589814 17372656 16598258 18521948 22491943 25659376 30467170 25830018 25348373 22936249 24495485 25474469 28004739
21719571 17994087 18057021 20920257 23761445 25244985 26483478 17510324 20966253 24438588 15632085 23185243 17550304 25315136 26108605 8201616 10731132 12537572 12537569 8216259 7589814 17372656 16598258 18521948 22491943 25659376 30467170 25830018 25348373 22936249 24495485 25474469 28004739
EMBL
KQ459595
KPI95733.1
KQ461192
KPJ07027.1
GDQN01003710
JAT87344.1
+ More
GDQN01010062 GDQN01009104 GDQN01007779 JAT80992.1 JAT81950.1 JAT83275.1 NWSH01000384 PCG76793.1 RSAL01000001 RVE55153.1 BABH01026599 BABH01026600 JTDY01002788 KOB70729.1 FX985586 BBF97919.1 KQ979851 KYN18844.1 GL446181 EFN88334.1 GBYB01005532 JAG75299.1 KK107522 EZA49318.1 KQ978023 KYM97873.1 ADTU01013250 KQ982696 KYQ51980.1 KQ976401 KYM92527.1 KQ759817 OAD62770.1 GFDF01000172 JAV13912.1 NEVH01015298 PNF27310.1 PNF27312.1 GL888182 EGI65665.1 GGFK01004356 MBW37677.1 GANO01001623 JAB58248.1 CH933809 KRG05987.1 KRG05988.1 EDW18118.1 GGFK01004141 MBW37462.1 ADMH02000614 ETN65622.1 AXCN02001655 GFDL01006559 JAV28486.1 GFDL01006669 JAV28376.1 JXJN01009740 GFDL01006514 JAV28531.1 GFDL01006567 JAV28478.1 GFDL01006509 JAV28536.1 CP012525 ALC42993.1 CH916366 EDV96408.1 CH940647 KRF84347.1 KRF84348.1 JXUM01026809 JXUM01026810 KQ560768 KXJ80961.1 EDW69377.2 CH477814 EAT35966.1 DS232438 EDS41741.1 KQ435922 KOX68473.1 CH902618 KPU78069.1 KPU78070.1 ATLV01009818 KE524548 KFB35473.1 EDV39500.2 OUUW01000012 SPP87269.1 CH379070 EAL29947.3 KRT08507.1 KRT08508.1 CH964095 EDW79294.1 GFDG01001150 JAV17649.1 KA644801 AFP59430.1 CM000159 EDW92821.2 KRK00729.1 KRK00730.1 KZ288198 PBC33660.1 BT133300 AFD10747.1 JRES01000356 KNC31972.1 U07596 AE014296 AY069044 L13070 AJ000879 CH954178 KQS43058.1 EDV50147.2 KQS43059.1 KQS43060.1 GBXI01013699 GBXI01010609 JAD00593.1 JAD03683.1 GBXI01015820 JAC98471.1 GAKP01010599 JAC48353.1 GAKP01010593 JAC48359.1 CM002912 KMY96796.1 GAMC01020721 GAMC01020720 JAB85834.1 KMY96793.1 GDHF01018788 GDHF01014275 JAI33526.1 JAI38039.1 GDHF01020202 JAI32112.1 KMY96794.1 KMY96795.1 CH480817 EDW50033.1 APCN01002381 GBBI01000313 JAC18399.1 GEDC01025847 JAS11451.1 GEZM01099418 GEZM01099415 GEZM01099412 GEZM01099410 GEZM01099409 JAV53330.1
GDQN01010062 GDQN01009104 GDQN01007779 JAT80992.1 JAT81950.1 JAT83275.1 NWSH01000384 PCG76793.1 RSAL01000001 RVE55153.1 BABH01026599 BABH01026600 JTDY01002788 KOB70729.1 FX985586 BBF97919.1 KQ979851 KYN18844.1 GL446181 EFN88334.1 GBYB01005532 JAG75299.1 KK107522 EZA49318.1 KQ978023 KYM97873.1 ADTU01013250 KQ982696 KYQ51980.1 KQ976401 KYM92527.1 KQ759817 OAD62770.1 GFDF01000172 JAV13912.1 NEVH01015298 PNF27310.1 PNF27312.1 GL888182 EGI65665.1 GGFK01004356 MBW37677.1 GANO01001623 JAB58248.1 CH933809 KRG05987.1 KRG05988.1 EDW18118.1 GGFK01004141 MBW37462.1 ADMH02000614 ETN65622.1 AXCN02001655 GFDL01006559 JAV28486.1 GFDL01006669 JAV28376.1 JXJN01009740 GFDL01006514 JAV28531.1 GFDL01006567 JAV28478.1 GFDL01006509 JAV28536.1 CP012525 ALC42993.1 CH916366 EDV96408.1 CH940647 KRF84347.1 KRF84348.1 JXUM01026809 JXUM01026810 KQ560768 KXJ80961.1 EDW69377.2 CH477814 EAT35966.1 DS232438 EDS41741.1 KQ435922 KOX68473.1 CH902618 KPU78069.1 KPU78070.1 ATLV01009818 KE524548 KFB35473.1 EDV39500.2 OUUW01000012 SPP87269.1 CH379070 EAL29947.3 KRT08507.1 KRT08508.1 CH964095 EDW79294.1 GFDG01001150 JAV17649.1 KA644801 AFP59430.1 CM000159 EDW92821.2 KRK00729.1 KRK00730.1 KZ288198 PBC33660.1 BT133300 AFD10747.1 JRES01000356 KNC31972.1 U07596 AE014296 AY069044 L13070 AJ000879 CH954178 KQS43058.1 EDV50147.2 KQS43059.1 KQS43060.1 GBXI01013699 GBXI01010609 JAD00593.1 JAD03683.1 GBXI01015820 JAC98471.1 GAKP01010599 JAC48353.1 GAKP01010593 JAC48359.1 CM002912 KMY96796.1 GAMC01020721 GAMC01020720 JAB85834.1 KMY96793.1 GDHF01018788 GDHF01014275 JAI33526.1 JAI38039.1 GDHF01020202 JAI32112.1 KMY96794.1 KMY96795.1 CH480817 EDW50033.1 APCN01002381 GBBI01000313 JAC18399.1 GEDC01025847 JAS11451.1 GEZM01099418 GEZM01099415 GEZM01099412 GEZM01099410 GEZM01099409 JAV53330.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000283053
UP000005204
UP000037510
+ More
UP000078492 UP000008237 UP000053097 UP000078542 UP000005203 UP000005205 UP000075809 UP000078540 UP000069272 UP000235965 UP000007755 UP000009192 UP000000673 UP000075886 UP000078200 UP000076408 UP000092443 UP000092460 UP000092553 UP000001070 UP000008792 UP000069940 UP000249989 UP000075881 UP000008820 UP000002320 UP000053105 UP000075902 UP000075903 UP000075882 UP000092445 UP000075880 UP000075920 UP000007801 UP000030765 UP000091820 UP000268350 UP000075885 UP000001819 UP000007798 UP000002282 UP000095301 UP000242457 UP000037069 UP000000803 UP000008711 UP000192221 UP000001292 UP000095300 UP000075900 UP000075840
UP000078492 UP000008237 UP000053097 UP000078542 UP000005203 UP000005205 UP000075809 UP000078540 UP000069272 UP000235965 UP000007755 UP000009192 UP000000673 UP000075886 UP000078200 UP000076408 UP000092443 UP000092460 UP000092553 UP000001070 UP000008792 UP000069940 UP000249989 UP000075881 UP000008820 UP000002320 UP000053105 UP000075902 UP000075903 UP000075882 UP000092445 UP000075880 UP000075920 UP000007801 UP000030765 UP000091820 UP000268350 UP000075885 UP000001819 UP000007798 UP000002282 UP000095301 UP000242457 UP000037069 UP000000803 UP000008711 UP000192221 UP000001292 UP000095300 UP000075900 UP000075840
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
A0A194PRZ8
A0A0N1INH8
A0A1E1WK14
A0A1E1W8K0
A0A2A4JZ64
A0A3S2MAG9
+ More
H9JK06 A0A0L7L631 A0A348G646 A0A151J6J8 E2B7F9 A0A0C9QXW7 A0A026VZT4 A0A195CCQ9 A0A088AVI4 A0A158NEW1 A0A151WVP6 A0A195BVC0 A0A310ST08 A0A1L8E5X6 A0A182FKP9 A0A2J7QFE4 A0A2J7QFE9 F4WJ87 A0A2M4AA58 U5EW26 A0A0Q9XL00 B4L0P1 A0A2M4A9P6 W5JML8 A0A182QYK2 A0A1Q3FLX3 A0A1Q3FLL4 A0A1A9V471 A0A182YL50 A0A1A9YEG4 A0A1B0B7X8 A0A1Q3FLT0 A0A1Q3FLT2 A0A1Q3FM21 A0A0M3QVU1 B4IXH1 A0A0Q9WHM7 A0A182H660 B4LG69 A0A182KAK6 A0A1S4FUJ9 Q16NP7 B0X747 A0A0M8ZS21 A0A182TTN5 A0A182VJX7 A0A182L4J8 A0A1A9ZS32 A0A182IYV6 A0A182WED5 A0A0P8ZU70 A0A084VBX9 B3M521 A0A1A9WLI6 A0A3B0KL26 A0A182PEJ1 Q29EW2 A0A0R3P5Y2 B4N3T9 A0A1L8EGC1 T1P833 B4PD73 A0A0R1DZQ6 A0A1I8N9X0 A0A1I8N9W3 A0A2A3EPN8 H8F4R0 A0A0L0CIE1 Q23979 Q23979-2 Q23979-3 A0A0Q5UH73 B3NB94 A0A0Q5U5L1 A0A0A1WZ38 A0A0A1WHX9 A0A034W3K4 A0A034W101 A0A0J9RLM0 W8B7N0 A0A0J9RLH4 A0A0K8V3T3 A0A0K8UZK9 A0A0J9RN04 A0A1W4W7N5 B4HVR8 A0A1W4VV78 A0A1W4W748 A0A1I8P1M8 A0A182R4F7 A0A182HH34 A0A023FBM3 A0A1B6CDD8 A0A1Y1JZT2
H9JK06 A0A0L7L631 A0A348G646 A0A151J6J8 E2B7F9 A0A0C9QXW7 A0A026VZT4 A0A195CCQ9 A0A088AVI4 A0A158NEW1 A0A151WVP6 A0A195BVC0 A0A310ST08 A0A1L8E5X6 A0A182FKP9 A0A2J7QFE4 A0A2J7QFE9 F4WJ87 A0A2M4AA58 U5EW26 A0A0Q9XL00 B4L0P1 A0A2M4A9P6 W5JML8 A0A182QYK2 A0A1Q3FLX3 A0A1Q3FLL4 A0A1A9V471 A0A182YL50 A0A1A9YEG4 A0A1B0B7X8 A0A1Q3FLT0 A0A1Q3FLT2 A0A1Q3FM21 A0A0M3QVU1 B4IXH1 A0A0Q9WHM7 A0A182H660 B4LG69 A0A182KAK6 A0A1S4FUJ9 Q16NP7 B0X747 A0A0M8ZS21 A0A182TTN5 A0A182VJX7 A0A182L4J8 A0A1A9ZS32 A0A182IYV6 A0A182WED5 A0A0P8ZU70 A0A084VBX9 B3M521 A0A1A9WLI6 A0A3B0KL26 A0A182PEJ1 Q29EW2 A0A0R3P5Y2 B4N3T9 A0A1L8EGC1 T1P833 B4PD73 A0A0R1DZQ6 A0A1I8N9X0 A0A1I8N9W3 A0A2A3EPN8 H8F4R0 A0A0L0CIE1 Q23979 Q23979-2 Q23979-3 A0A0Q5UH73 B3NB94 A0A0Q5U5L1 A0A0A1WZ38 A0A0A1WHX9 A0A034W3K4 A0A034W101 A0A0J9RLM0 W8B7N0 A0A0J9RLH4 A0A0K8V3T3 A0A0K8UZK9 A0A0J9RN04 A0A1W4W7N5 B4HVR8 A0A1W4VV78 A0A1W4W748 A0A1I8P1M8 A0A182R4F7 A0A182HH34 A0A023FBM3 A0A1B6CDD8 A0A1Y1JZT2
PDB
4BYF
E-value=0,
Score=2224
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Protein shifts from the basolateral to apical domain in enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:22491943, PubMed:25659376). With evidence from 7 publications.
Cell cortex Protein shifts from the basolateral to apical domain in enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:22491943, PubMed:25659376). With evidence from 7 publications.
Cell membrane Protein shifts from the basolateral to apical domain in enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:22491943, PubMed:25659376). With evidence from 7 publications.
Cell cortex Protein shifts from the basolateral to apical domain in enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:22491943, PubMed:25659376). With evidence from 7 publications.
Cell membrane Protein shifts from the basolateral to apical domain in enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:22491943, PubMed:25659376). With evidence from 7 publications.
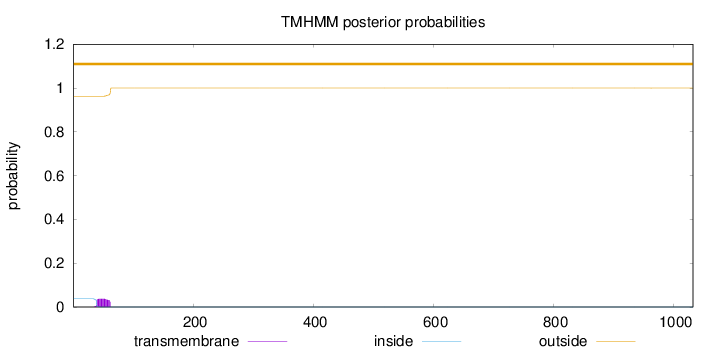
Length:
1032
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.825689999999999
Exp number, first 60 AAs:
0.76653
Total prob of N-in:
0.03733
outside
1 - 1032
Population Genetic Test Statistics
Pi
242.716803
Theta
160.850652
Tajima's D
0.892276
CLR
0.437625
CSRT
0.630968451577421
Interpretation
Uncertain
